Viruses of the Picornaviridae family of which enteroviruses (EVs) belong are non-enveloped and icosahedral viruses, measure 30–32 nm in diameter and present positive-sense, non-segmented RNA genome ranging in size from 6.7 to 10.1 kb containing a single long open reading frame. They infect humans and some animal species, and are taxonomically grouped into A-L Enterovirus species and A-C Rhinovirus species [Reference Zell1]. The classification of the EVs has been constantly changing with the discovery of new genetic sequences. The known bovine enteroviruses (BEV) belong to the species EV-E and -F; EV-E is subgrouped into four types and EV-F into six types. Details can be found in the Picornaviruses database [2].
Occasionally the virus causes serious and life-threatening diseases in humans and animals, but most EVs infections are subclinical [Reference Blas-Machado3, Reference Palacios and Oberste4]. In bovine, EVs have been isolated from cattle with a wide range of clinical signs including enteric infections (high morbidity, diarrhoea and moderate mortality), respiratory disease (cough, fever and dyspnoea), reproductive disorders and infertility [Reference Ze-Li5, Reference Zhang6]. EVs are important markers of faecal contamination by cattle, occurring more commonly in rural environments [Reference Blas-Machado7].
The first isolation of BEV occurred in the late 1950s [Reference McFerran8]. The first and only work performed with these viruses in Brazil dates back more than 40 years. This work described the detection of BEV in two different Brazilian states (São Paulo and Pernambuco) through serological screening [Reference Linhares9].
We present an important update of the presence of BEV in Brazil and also the first molecular characterisation of these viruses present in different Brazilian states.
From 15 July 2012 to 18 March 2016, 103 faecal samples were collected from the rectum of cattle (beef and dairy) in rural areas of the states of São Paulo, Minas Gerais, Goiás, Rio Grande do Sul, Paraná and Mato Grosso do Sul in Brazil. Of the studied animals, 36 (34.9%) had diarrhoea, 67 (65.0%) were aged <4 months and the majority (71 animals, 68.9%) were female. Seventy-four (71.8%) were raised under the feedlot system and 29 (28.1%) under free-range production conditions. Immediately after collection, the samples were kept in plastic bags at −4 °C until processing in the laboratory.
The RNA of samples was extracted using TRIzol™ Reagent (Invitrogen, USA). Reverse transcription was performed using the ImProm-II™ Reverse Transcription System (Promega, USA) and random primers (Invitrogen, USA), according to the manufacturer's instructions. Polymerase chain reaction (PCR) targeting a 183-bp fragment containing a part of the 5′-terminal portion of BEV [Reference Ley, Higgins and Fayer10] was done with a GoTaq™ Colorless Master Mix (Promega, USA), following the manufacturer's protocol, using the bovine β-actin gene as internal control [Reference Renshaw, Ray and Dubovi11]. The amplified products were gel extracted and sequenced directly on both strands with the same primers used in the PCR, in an automated ABI 3730 DNA Analyzer (Applied Biosystems, USA).
The results showed that 15 (14.5%) of the samples collected were positive for BEV including both calves and adult animals; five animals had diarrhoea, 11 were females and the majority of the positive samples were from dairy cattle (n = 13). Ten animals were from the state of São Paulo, four were from the state of Minas Gerais and one was from the state of Goiás.
Out of all PCR-positive samples, seven were selected and submitted to nucleotide sequencing because they represented the different cattle farms with BEV-PCR-positive results. However, only four sequences showed quality for subsequent analyses (two from state of São Paulo, one from state of Minas Gerais and one from state of Goiás). These four sequences shared >92.2% nt sequence identity when compared each other, 81.1%–86.6% nt identity when compared with Enterovirus E samples and 86.1%–92.8% nt identity when compared with Enterovirus F samples. Phylogenetic analysis of the partial sequences clustered these BEV Brazilian samples into the Enterovirus F clade (Fig. 1). Our results presented, after more than four decades, the occurrence of BEV in Brazil and the first phylogenetic characterisation of these viruses found in Brazilian cattle herds.
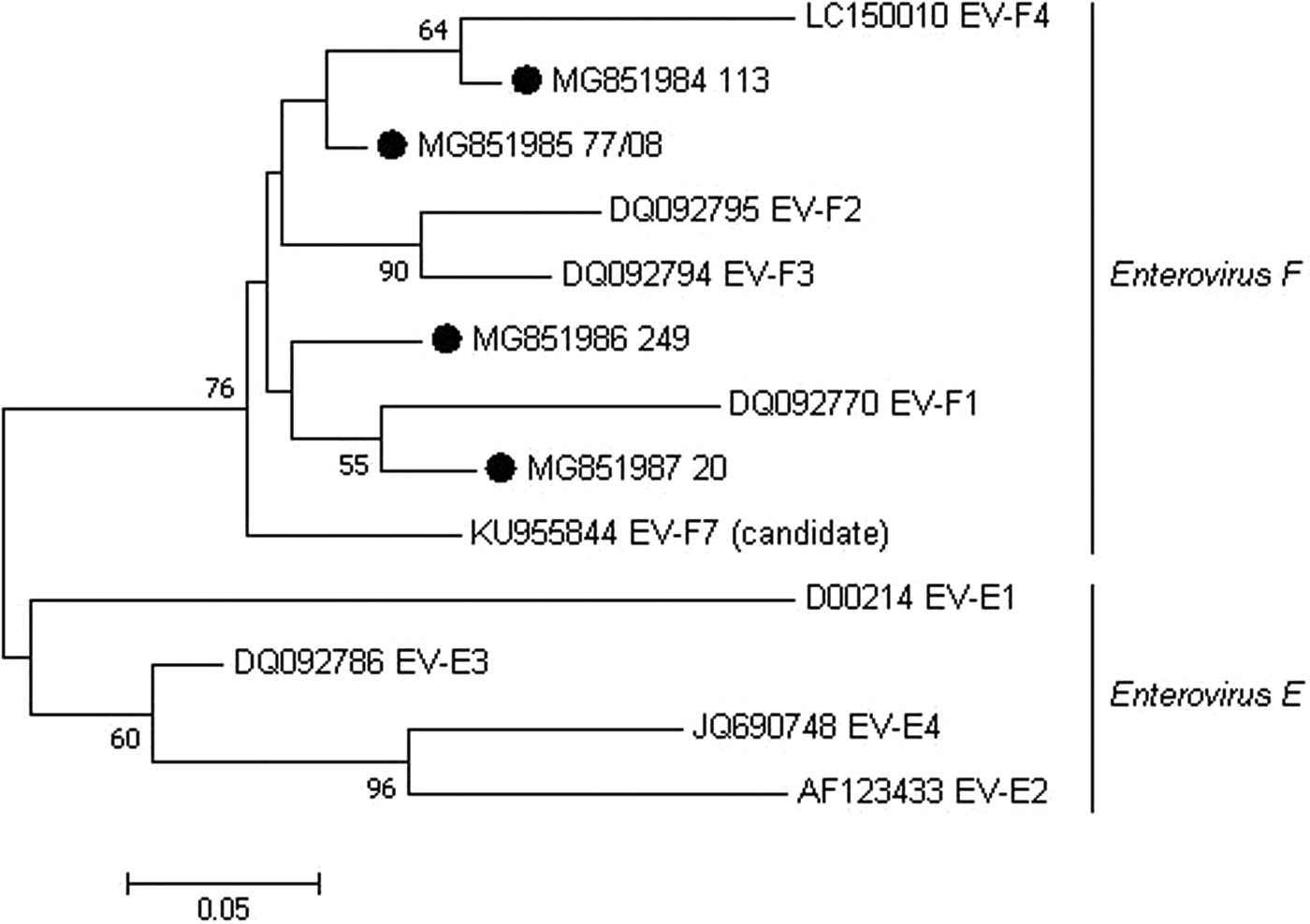
Fig. 1. Maximum-likelihood unrooted phylogenetic relationships using a 183 bp-sequence of the 5′-terminal portion of BEV. Bootstrap values higher than 50% for 1000 pseudo-replicates are showed at the nodes. The sequences obtained in the present study are labelled with a filled circle. GenBank accession numbers and representative samples of the EV subgroups E and F are shown on the tree. The scale bar represents the phylogenetic distance among sequences.
BEV-infected cattle shed large amounts of virus in their faeces leading to environmental contamination, these animals constantly present mild to severe respiratory complications, varying degrees of diarrhoea and infertility [Reference Zhang6].
Most of the samples used in this study were also used to investigate other viruses that cause gastroenteritis in cattle, and one of the samples was positive for both BEV and bovine Astrovirus [Reference Candido12]; two other samples were positive for both BEV and bovine Kobuvirus [Reference Candido13] and two samples were positive for both BEV and bovine Picobirnavirus [Reference Navarro14]. It is interesting to note that in all cases of coinfection the animals were less than 8 months old. Some authors also reported the presence of different viruses in animal samples diagnosed with BEV [Reference Zhang6, Reference Zhu15].
In this work we found BEV in animals without apparent diarrhoea and in adult animals, thus we cannot affirm that the viral strains detected in this study trigger diarrhoea in infected animals or are prevalent in younger animals.
This study is an important update of the presence of BEV in different Brazilian regions and describes the first partial molecular characterisation of these viruses in Brazil. Although it is not yet possible to define the degree of risk of the presence of the virus in the confirmed regions, there are several reports of serious problems involving BEV infection around the world, from gastroenteritis, respiratory problems to abortions [Reference Ze-Li5, Reference Zhang6], which could lead to incalculable losses to the cattle farming sector.
Future studies using specific primers for the verification of BEV-E or even the complete genome of BEVs are required for the understanding of the diversity of strains and subgroups of these viruses in Brazil. In addition, the experimental infection of gnotobiotic cattle using BEV strains circulating in the different Brazilian states is necessary for a better understanding of the severity of the circulating viral strains in the country. Our results contribute to a better understanding of the distribution and characterisation of viral agents potentially associated with enteric, respiratory and reproductive infections in cattle.
Author ORCID
Marcelo Candido, 0000-0001-9067-4065.
Financial support
This study was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, nos. 2006/52060-3 and 2012/18441-0) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, no. 472509/2010-1).
Conflict of interest
None.



