Introduction
Noroviruses are the leading causative agent of acute gastroenteritis (AGE), infecting peoples of all age groups. These viruses are responsible for about a fifth (18%) of all AGE cases worldwide, leading to about 685 million illnesses, 210 000 deaths and 15 million disability-adjusted life years annually [Reference Lopman1]. In recent years, many countries have been experiencing an increase in AGE outbreaks associated with noroviruses, causing significant disease burden [Reference Zhou2, Reference Bányai3]. Currently, there is no vaccine or specific treatment against noroviruses.
Noroviruses are single-stranded, positive-sense RNA viruses with a genome of 7.5–7.7 kb, being divided into 10 genogroups (GI–GX) [Reference Chhabra4]. Noroviruses use glycans of histo-blood group antigen (HBGA) for attachment to their target cells. These glycans are mainly controlled by the fucosyltransferase 2 (FUT2) in secretors, FUT3 (Lewis) and the ABH genes families. Individuals with FUT2 enzyme expressions are secretor-positive, while those without are secretor-negative and they do not express ABH HBGAs on mucosal surfaces [Reference Nordgren and Svensson5–Reference Tan and Jiang7]. Various HBGAs binding profiles of norovirus-HBGA interactions are typical among different noroviruses genotypes. The GII.6 noroviruses are consistently associated with infections among young children and the elderly [Reference Parra and Green8, Reference Bruggink, Moselen and Marshall9]. Genogroups II.6 were the second most common genotype in active AGE surveillance from 2011 to 2015 in the USA [Reference Cannon10]. Studies revealed it tended to infect FUT2 secretor status [Reference Currier11] and was significantly less likely to have symptoms in non-secretors [Reference Sharma12]. Another study analysing sequential outbreaks caused by GII.2 and GII.6 noroviruses in institutional settings revealed that 21% of residents who shed GII.6 noroviruses and developed antibodies against GII.6, were secretor-negative [Reference Karangwa13]. Genogroups II.6 noroviruses consist of three clusters, differing in amino acid residues (5.2–9.2%) from each other that can be detected after 40 years [Reference Parra14]. Herd immunity to noroviruses is complicated due to its diversity. Thus, the roles of HBGA and immunity statuses of individuals on GII.6 norovirus infections require further investigations.
Here, we reported an AGE outbreak caused by GII.6/b noroviruses in November 2018 at a hospital in Jinshan District, Shanghai, China. Studies on molecular epidemiology, host susceptibility and serological roles were performed to understand the GII.6/b noroviruses as an AGE outbreak agent.
Materials and methods
Outbreak investigation and samples collection
An AGE outbreak occurred from 23 to 29 November 2018 at a specialised hospital in Jinshan District, Shanghai, China. Symptomatic cases were defined as individuals who experienced symptoms of at least one episode of vomiting and diarrhoea from 23 November. Confirmed cases were those whose stool specimen or rectal swab was positive for norovirus. Asymptomatic cases were defined as those who did not experience symptoms of vomiting or diarrhoea but whose stool specimen or rectal swab was positive for norovirus [Reference Zheng15].
The affected hospital contained four care units (CU) with 439 inpatients and 111 staff, including 24 doctors, 75 nurses, seven canteen food workers and five logistic workers. The four CU were on different floors. Each of the CU had 12 wards, one public toilet, a public bathroom and a common activity room for all inpatients. The wards were in proximity but without a bathroom.
Twenty-three stool samples were collected from symptomatic individuals (22 inpatients and one doctor). Additionally, 13 stool samples from asymptomatic individuals were collected from a patient who shared the ward with the index case, seven canteen food workers and five from logistic workers. Continuous collections and norovirus detection of stool samples were performed for those staff with positive results on 3, 7, 10, 17 and 24 December. Out of 25 saliva samples, 20 from symptomatic and five asymptomatic individuals were collected. Twenty-two paired serum samples from symptomatic individuals at the acute phase (within 1 week after onset date) and convalescent phase (about 4 weeks after the onset date). Furthermore, two paired serum samples from asymptomatic individuals at the parallel time were also collected.
Pathogens detection, cloning and sequencing of the norovirus P domain
Norovirus was detected using a one-step real-time polymerase chain reaction (PCR) with GI and GII primers [Reference Wolf16] and primer COG2F and GIISKR targeting at NS terminus in ORF 2 were further used for further genotyping [Reference Kojima17]. Positive PCR products were sequenced and norovirus genotype was determined by genotyping tool [Reference Kroneman18].
The P domain of the outbreak norovirus strain (Jinshan 06) was amplified using primers GII.6-F: 5′-CGGATCCTCAAAGACTAAGCCCTTTACA-3′ and GII.6-R: 5′-GCATGCGCCGCTTAGCAAAAGCAATCGCCACGCAATCGCAATCGCATTGAACTCTTCTG-3′. The reverse primer was linked by a cysteine-containing peptide (P-CDCRGDCFC) to C-terminus, as described previously [Reference Tan and Jiang19]. The P domain sequences were cloned into T-vectors (Thermo Fisher, US). Phylogenetic trees were constructed using the neighbour-joining method (MEGA 6.0).
HBGA phenotyping of saliva samples
HBGA phenotyping of saliva samples was performed as described earlier [Reference Dai20]. Boiled saliva at a dilution of 1:1000 was coated on 96-well ELISA plates (high binding, Costar, Corning, CN). After blocking with PBS – 5% non-fat milk, monoclonal antibodies (anti – A, B and H, Santa Cruz, CA, USA; Lea, Leb, Lex and Ley, Signet Laboratories Inc., Dedham, MA, USA) at a dilution of 1:300 were added to detect HBGAs. The signals were read using a TMB substrate kit. The cut-off of a positive signal was set at 0.2 as an approximate value of the mean of the background/blank wells adding a triple standard deviation. As quality controls, well-characterised positive and negative saliva samples were added in each plate.
HBGA binding profiles of GII.6 norovirus
Saliva-based HBGA-P protein-binding assay was performed as described in Ref. [Reference Zhang21]. Well-characterised boiled saliva samples [Reference Tang22] at 1:1000 dilution were coated onto plates overnight at 4 °C. After blocking with 5% PBS-non-fat milk, P proteins at a concentration of 5μg/ml of the outbreak strain were added and incubated at 37 °C for 1 h. Then our in-house made mice anti-GII.6 norovirus (dilution, 1:3000) sera were added, followed with HRP-conjugated goat anti-mice IgG at 1:5000. The cut-off of a positive signal was set at 0.2.
GII.6 norovirus specific IgG antibody titres in paired sera
Plates were coated with 0.5 μg/ml diluted P protein of the outbreak strain. After blocking with 5% non-fat milk in PBS, patient serum samples at serial twofold dilution (1:500–1:128 000) were added. The bound norovirus antibodies were then detected by horseradish peroxidase (HRP)-conjugated goat anti-human IgG (dilution, 1:3000). A cut-off was also set at OD450 = 0.2 and a titre of <500 was assigned to a value of 250. Seroconversion was defined as ≥⩾fourfold increase in titre of the convalescent-phase serum sample compared with that of the acute-phase serum sample.
Blockade of the binding of P protein to HBGAs in saliva by paired sera
A well-characterised saliva sample with HBGA phenotypes of B, Leb and Ley, which showed good stable binding to the P protein of GII.6 norovirus, was used in HBGA blockade assays. The blockade assay was HBGA binding assay (see above) with an extra blockade step. The P proteins (5 μg/ml) of GII.6 norovirus were pre-incubated with paired sera in twofold serial dilution (1:25–1:3200) for 1 h at 37 °C and added to the coated saliva. The positive control consisted of P proteins without pre-incubation with a serum sample and showed ODs of 1.0 ± 0.3 [Reference Reeck23]. The 50% blocking titre (BT50) defined as the maximal dilutions (folds) of a serum sample that showed at least 50% blockade in OD compared with the positive control. Serum samples with a BT50 <25 were assigned a value of 12.5 [Reference Reeck23].
Statistical analysis
The correlation between IgG and serum blockade titres was determined using the Pearson correlation coefficient test by SPSS version 22.0 (SPSS Inc., Chicago, IL, USA).
Results
Descriptive epidemiology
A total of 27 AGE cases (26 inpatients and one doctor) in two care units met case criteria. All cases were males with a median age of 45.1, ranging from 27 to 65 years. The main symptoms included vomiting (77.1%) and diarrhoea (44.4%) (Table 1).
Table 1. Basic demographics, symptoms, stool tests and HBGA phenotypes of symptomatic and asymptomatic cases
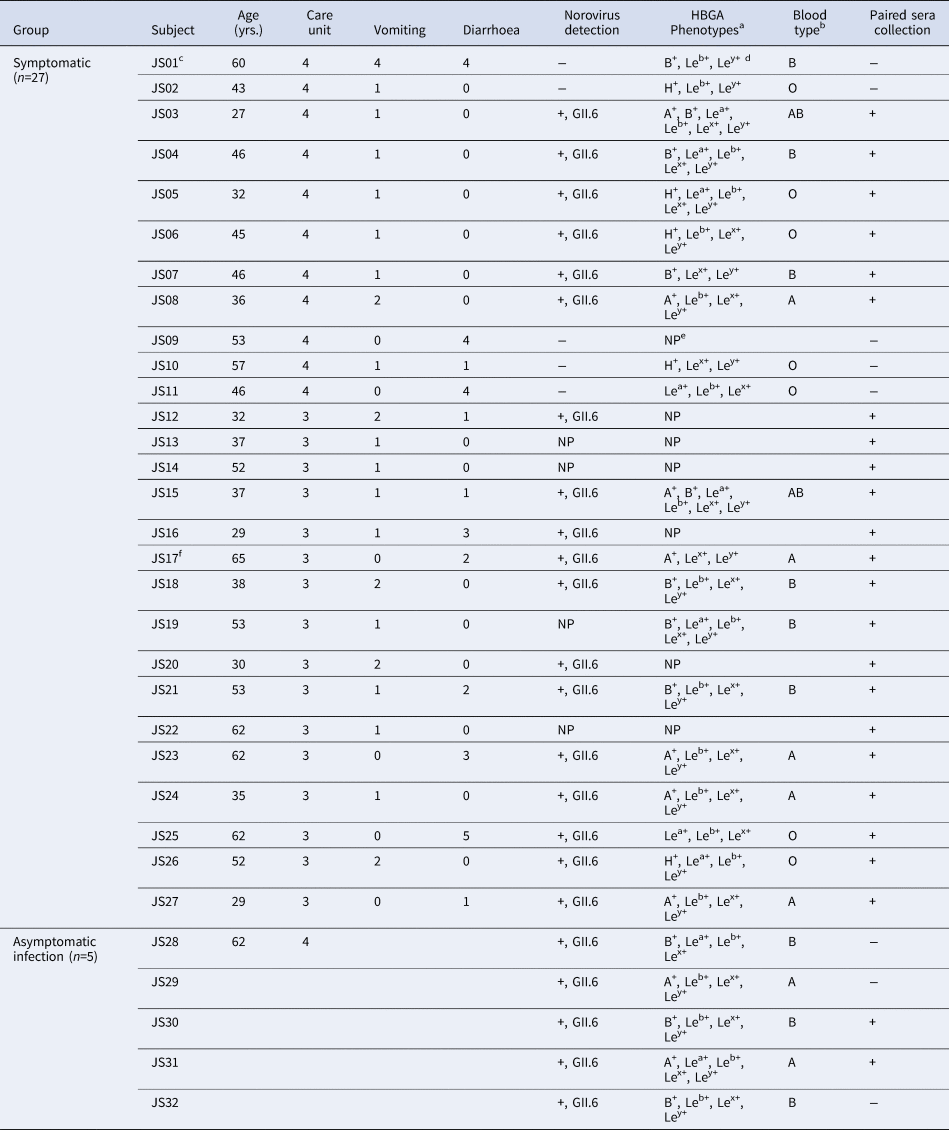
a The subjects with HBGA phenotype indicating collection of saliva.
b All individuals were detected as secretor with HBGA phenotyping.
c The first case.
d Le = Lewis.
e NP, not perform without collecting saliva samples.
f Doctor.
The index case was a 60-year-old male in CU4 with sudden onset symptoms of diarrhoea at 4:00 a.m. on 23 November. He experienced four episodes of watery diarrhoea and four episodes of vomiting in the public toilet room of CU4. Eleven cases were later identified in the same unit, with an attack rate of 9.02% (11/122) from 23 to 25 November. The first case in CU3 was a 30-year-old male on 26 November and had two episodes of watery diarrhoea and one episode of vomiting from 9:30 p.m to 11:00 p.m in the public toilet room of CU3. Sixteen cases were later identified in CU3 with an attack rate of 11.68% (16/137) between 26 and 29 November (Fig. 1).
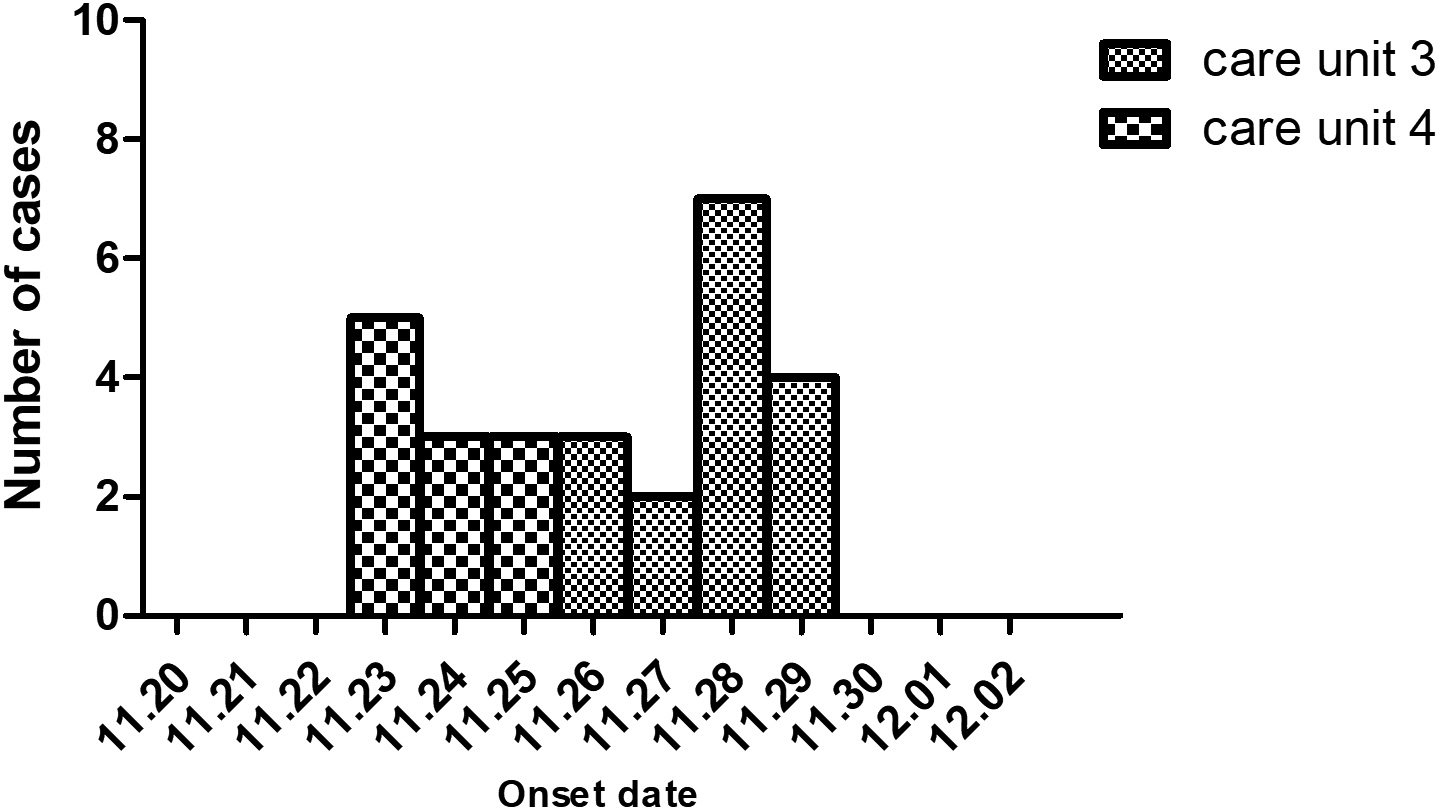
Fig. 1. The epidemic curve of the studied acute gastroenteritis outbreak by date of onset in a hospital of Jinshan in November 2018. Column chart indicating the numbers of acute gastroenteritis cases (Y-axis) in the two care units. X-axis shows dates.
Since this was a particular hospital for mental health patients, movement, food and drinking water were strictly regulated, excluding pathogens from other resources out of the hospital. Drinking water was supplied in the whole hospital. The fact that only CU4 and CU3 patients were infected indicated that water-borne was unlikely the cause of the outbreak.
Norovirus detection and phylogenetic analysis
Eighteen (78.26%, 17 inpatients and one doctor) out of 23 symptomatic cases and five (38.46%, two food workers, two logistic workers and one inpatient) out of 13 asymptomatic individuals were positive with GII.6 norovirus. The 23 sequences showed 99.7–100% nucleotide identity, indicating that this was a GII.6 norovirus outbreak (Fig. 2a). Further analysis of the P domain sequence revealed that the outbreak strain belonged to the GII.6/b cluster, sharing 99.3% homology with Groningen strain (LN854568) in the Netherlands (Fig. 2b). One confirmed symptomatic case and five (three males and two females) asymptomatic cases were found among the staff, including one doctor, two food workers and two logistic workers. No previous gastrointestinal illness was reported from all asymptomatic cases. Notably, two food handlers, who were positive for GII.6 norovirus without symptoms, were highly suspected as the source of the outbreak.
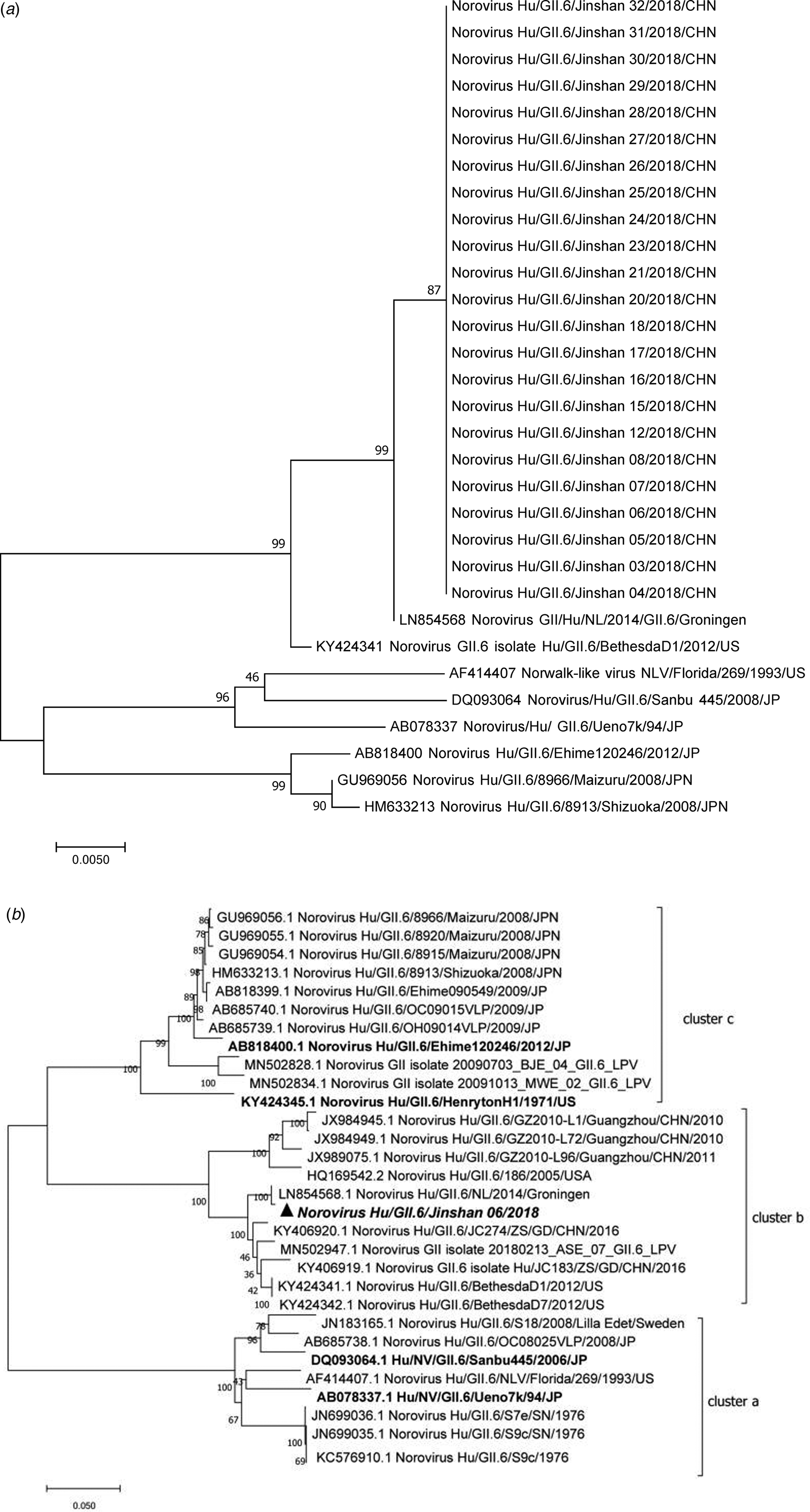
Fig. 2. Phylogenetic tree analyses based on nucleotide sequences encoding capsid NS terminus (387 bp fragment) (a) and P domain (b) (978 bp fragment, corresponding to the nucleotide 661–1638 of the VP1 encoding gene) of GII.6 noroviruses. The sequences isolated were marked with a triangle and the tree made with reference strains using the neighbour-joining method. The bootstrap values generated from 1000 replicates are shown at each node. GII.6 strains were classified into three distinct clusters a–c.
Among the confirmed cases, the doctor had diarrhoea at 2:00 a.m. on 28 November. He was in charge of CU3 and took on night shifts in CU4 and, thus, could have been the virus carrier leading to the second wave of outbreak. A high norovirus positive rate at 33.3% (4/12) from asymptomatic food and logistic workers was observed, who had high opportunities to spread the viruses. Furthermore, we noted that two infected staff (the doctor and one logistic worker) shed viruses for at least 19 days (until 17 December). This viral shedding might have contributed to spreading further and played a role in norovirus transmission.
HBGA-binding interfaces and the surrounding regions of GII.6 clusters
Although the sequence alignment by using MegAlign program of DNAstar 7.1 of different GII.6 clusters revealed highly conserved amino acids in HBGA-binding interfaces and the surrounding regions (Fig. 3), some variations were still observed. Compared with cluster a, one residue deletion in region 1 (A/-345) was seen in the HBGA-binding interfaces in clusters b and c, and a D–N mutation in the surrounding regions was also found in clusters b and c. Besides, one N/D/T/G390 mutation was also observed in region 2 among different clusters.
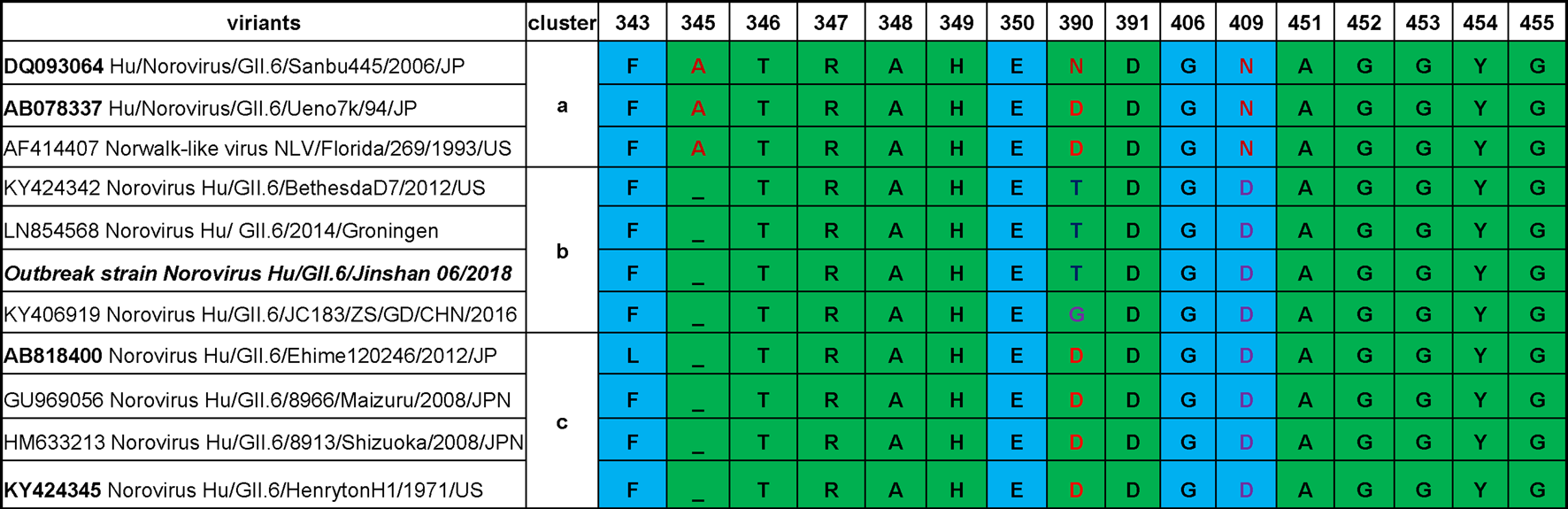
Fig. 3. Comparisons of P domain sequences that built the HBGA binding interfaces of GII.6 noroviruses and the neighbour regions. The representative sequences of GII.6 noroviruses are selected from different major phylogenetic clusters. The outbreak strain is shown with bold fonts. The sequences are aligned using MegAlign program (DNAstar 7.1). Amino acids in green are associated with HBGA binding, while those in light blue are related to the binding specificity based on previous studies [Reference Tan24, Reference Tan25].
HBGA phenotyping of saliva samples
The HBGA phenotypes of the saliva samples from 20 symptomatic and five asymptomatic infections were determined. Secretors were characterised by antigen content: H and A antigens for phenotype A; H, A and B antigens for the AB phenotype; H and B antigens for phenotype B; and H antigen for phenotype O. The phenotype of the non-secretors was characterised by low secretion of ABH antigens in saliva [Reference Shirato26]. All of them were identified as secretors, including seven A, nine B, two AB and seven O blood types (Table 1).
HBGA binding spectrum of the epidemic GII.6 variant
The results showed GII.6 P proteins bound saliva of type A, B, AB and O secretors. The binding signals were highest for type B and moderate for type A, AB and O saliva samples. However, there were no positive signals to the saliva of non-secretors (Fig. 4). The results were consistent with the observed HBGA-related host susceptibility.
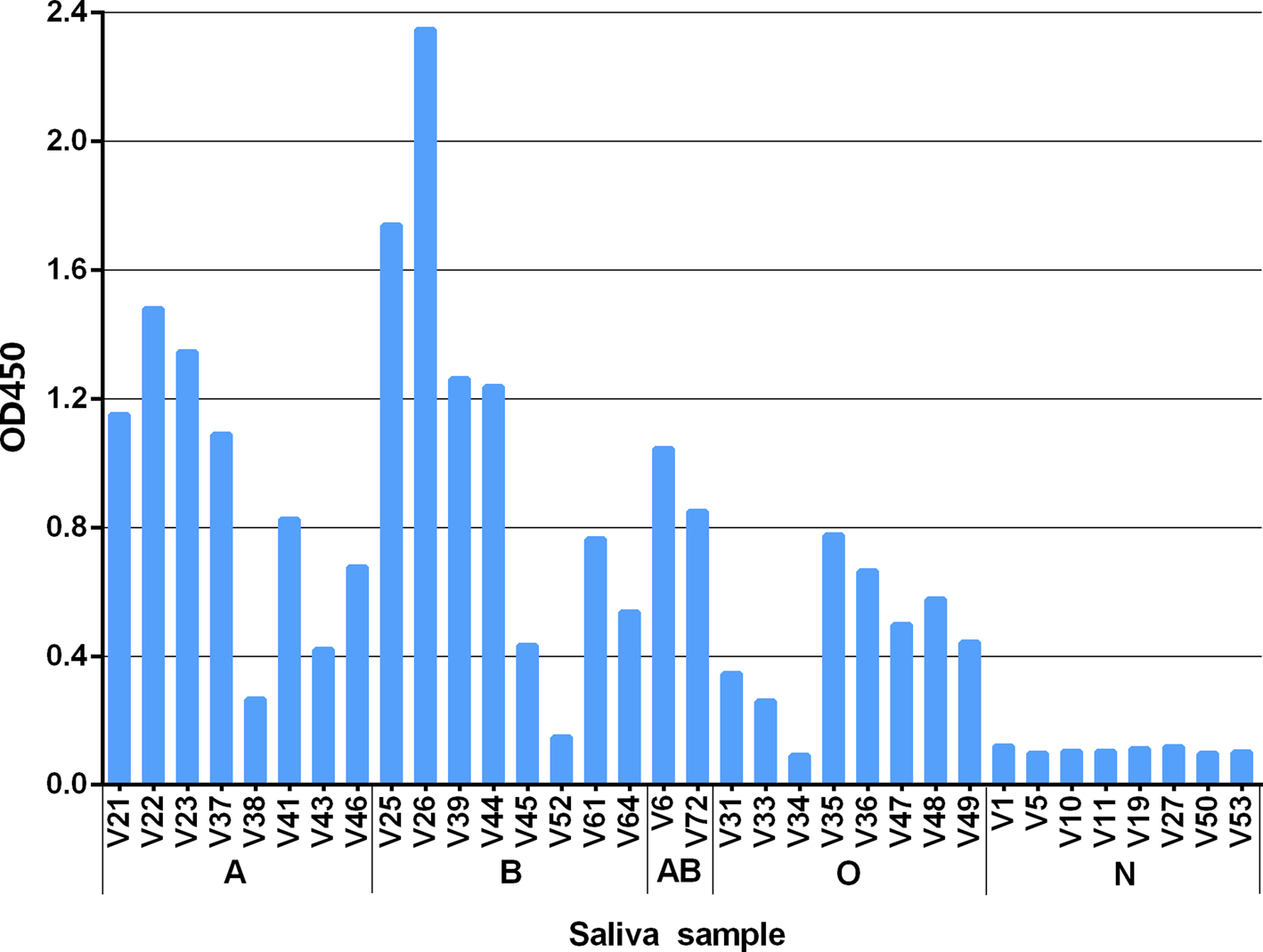
Fig. 4. Recombinant P protein of the studied GII.6/Jinshan 06 strain binding to well-characterised saliva samples representing various blood types. The binding signals are shown in optical density (Y-axis), while the well-characterised saliva (V as abbreviation for volunteers) [Reference Tang22] samples are shown in X-axis. A, B, O and N stands for blood type A, B, O and non-secretor, respectively.
Serum IgG titres of outbreak patients and blockade titres specific to GII.6 norovirus
The paired serum samples from both acute and convalescent phases were analysed for their GII.6 norovirus-specific IgG titres. Out of the 24 acute phase sera of cases, 20 (83.3%) possessed relatively high IgG titres of between 1000 and 8000. The IgG of the symptomatic cases showed higher titres in convalescent-phase sera, among which 19 showed ≥fourfolds increase (Fig. 5a). Two paired serum samples of asymptomatic cases did not show seroconversion.
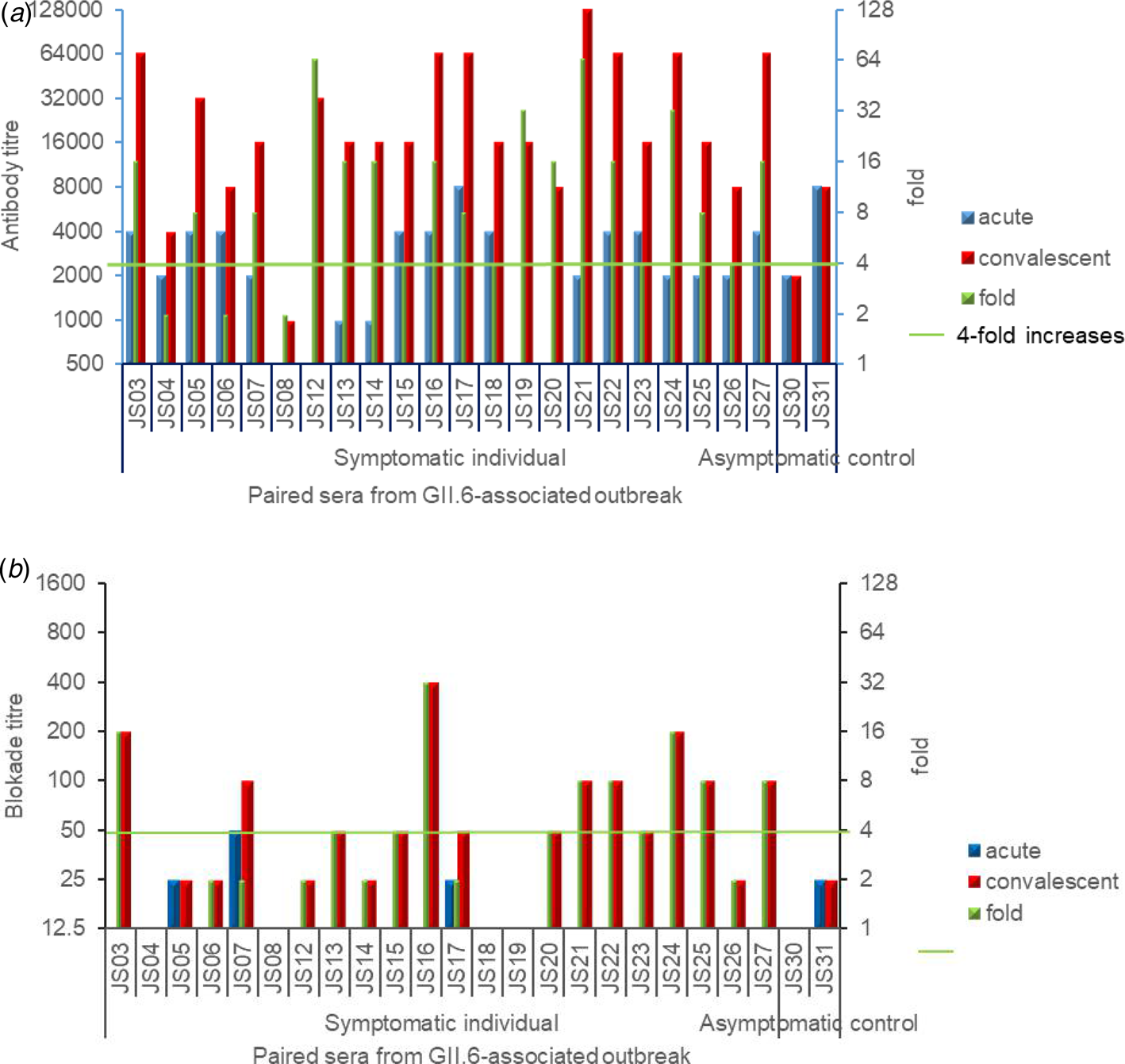
Fig. 5. (a) and (b): GII.6-specific IgG titres (a) and blockade titres (b) against NoV GII.6 P domain binding to HBGAs of the pared human sera collected from the outbreak. Blue (acute phase sera) and red (convalescent sera) columns show the IgG titres (a) or blockade titres (b), while different sera in indicating codes are shown in X-axis. The seroconversion between acute- and convalescent-phase sera were determined and shown. Green columns show the increased folds in GII.6 specific IgG titres or blocking titres by comparing their corresponding titres in the acute and convalescent sera.
With regards to blockade titres, only four out of the 24 individuals in the acute phase showed low detectable BT50 at 25 or 50. Among the symptomatic cases, 11 (11/22, 50%) seroconverted sera revealed ≥fourfolds blockade titre increase against GII.6 norovirus (Fig. 5b). Two paired serum samples of asymptomatic cases did not reveal a blockade seroconversion. These results indicated diverse immune responses to the GII.6 norovirus among individuals. No significant correlation was found between IgG titres and blockade titres in the acute phase (R = 0.264, P = 0.213), convalescent phase (R = 0.184, P = 0.390) and the fold of increase (R = 0.196, P = 0.358).
Discussion
Noroviruses are recognised as the leading cause of acute viral gastroenteritis worldwide in all age groups, due to their low infectious dose, environmental stability and diverse transmission modes [Reference Karst, Zhu and Goodfellow27]. Noroviruses-associated outbreaks or epidemics often occur in closed and semi-closed settings, such as large battleships, cruise ships, schools, hospitals and nursing homes [Reference Lian28].
Studies showed GII.6 noroviruses have widely circulated in many countries, such as in Korea, the USA, Peruvian, Australia and China [Reference Bruggink, Moselen and Marshall9, Reference Park29–Reference Saito31]. In Okayama, Japan, GII.6 norovirus-related infections accounted for 40.4% of all norovirus infection in non-hospitalised children with AGE, from 2008 to 2009 [Reference Chan-It32]. The virus was listed as the second most common genotype causing outbreaks from 2014 to 2015 among young children in the USA [Reference Cannon10]. As well as various other countries worldwide, molecular epidemiological studies in China also showed GII.6 noroviruses as commonly detected pathogens [Reference Guo33, Reference Luo34]. In this study, we reported an outbreak of gastroenteritis in a hospital in Jinshan, Shanghai, China, in November 2018. With the use of field epidemiology and serologic investigations, we showed a GII.6/b norovirus as the causative agent of the outbreak.
Numerous studies support an association between norovirus infection and the human HBGA phenotypes, which may be an essential factor affecting the prevalence of GII norovirus [Reference Zhang35–Reference Liu37]. GII.6 noroviruses consist of three clusters with some amino acid residues variations that can be detected after 40 years [Reference Parra14]. To further understand their prevalence in relation to HBGA binding profile, we analysed HBGA binding interfaces and the surrounding regions of different GII.6 clusters. Although these proteins were highly conserved, three amino acid residues mutations occurred among the clusters, which might lead to changes in HBGA binding patterns and affinities (Fig. 3). We noted that mutations occurred even in the same cluster. In a similar study involving two strains of cluster a, the A, B or O secretors bound the proteins, although one showed a weak bound to non-secretor salivas [Reference Shirato26]. Our analysis showed a D (AB078337) to N (DQ093064) mutation in residue 390 at the binding interface, which might have resulted in the observed HBGA binding variations. These differences among the GII.6 clusters highlight the importance of understanding the relationship between the genetic variations and the host susceptibility for specific norovirus strains. We noted that the previous studies on associations between HBGA binding profiles and GII.6 norovirus infection were focused on clusters a and c. The pathogen causing the outbreak of our study was a GII.6 cluster b norovirus. Our data showed that all symptomatic cases and asymptomatic infected individuals were A, B or O secretors, which was further supported by our saliva-based HBGA-P protein assays. The saliva binding results of our study showed a difference from the results indicated that GII.6 cluster c VLPs (AB818400) with the highest binding signals to type A and lowest signals to type O saliva samples. These variations may be due to the observed minor mutations in the HBGA binding interfaces.
Previous studies also showed that GII.6 norovirus recognises HBGAs of A, B and O secretors with some minor variations [Reference Shirato26, Reference Zheng38], suggesting that GII.6 noroviruses have a broad spectrum of target populations. Such studies also shed light on the host susceptibility, host range and epidemiology of GII.6 norovirus. Secretors represent 80–90% of the Chinese population [Reference Tan and Jiang39] and this could be a crucial factor contributing to the high prevalence of GII.6 in the country. Thus, further surveillance of norovirus GII.6 variants is needed to get in-depth knowledge.
Genotype-specific blocking antibodies are correlated with protection against norovirus infection and the magnitude of such blocking titres are related to protection from natural infection [Reference Malm40, Reference Atmar41]. In this study, 83.3% of the infected cases showed relatively high IgG titres between 1000 and 8000 at the acute phase and their sera did not reveal blockade antibody titres against GII.6 variant (Fig. 5). Only four infected individuals showed low blockade antibody titres between 25 and 50 (Fig. 3b), suggesting a low specific blockade antibody titre was correlated with infection at the beginning of the outbreak. Paired sera collected from norovirus outbreak gave valuable information for defining the correlation between protection and the magnitudes of pre-existing antibodies, as well as specific homologous immune response. It may also provide important data on cross immunity among different immunotypes, as newly recommended by Parra [Reference Parra14]. To elucidate antigenic relatedness among different norovirus immunotypes, characterisation on cross immunity in both GI and GII genotypes are ongoing in our laboratory. We found that half of the symptomatic patients showed 4–32-folds blocking titre increase against GII.6 variant in the outbreak and these paired sera could serve as precious materials to define cross reaction and cross blockade between GII.6 and other genotypes in the future.
Studies showed that symptomatic individuals, compared to asymptomatic, have greater immune system activation and suggested that symptoms may be immune-mediated in norovirus infection [Reference Newman42]. In the present study, we found that two asymptomatic cases did not seroconvert, which supported their observation.
Studies have shown that employees played an important role in the transmission during some norovirus outbreaks [Reference Chen43, Reference Xue44]. A study showed that the infection rate of asymptomatic employee was 17.0% from 49 norovirus outbreaks in Shanghai [Reference Wu45]. In our study, the norovirus positive rate of convenient samples from asymptomatic staffs were 33.3%, which was consistent with the previous findings [Reference Bok and Green46].
This study had some limitations. One was the relatively small size of infected individuals. A sampling of convenience may result in a selection bias that might further affect the accuracy of the study outcomes. Secondly, no saliva samples were collected from uninfected controls; as a result, the risk of secretor and non-secretor to GII.6 infection could not be evaluated.
In conclusion, the study represents an investigation on a gastroenteritis outbreak caused by GII.6/b norovirus, focusing on epidemiology, host susceptibility, host HBGA phenotypes and HBGA binding profiles of causative norovirus. A low specific blockade antibody in the population may play an important role at the beginning of the outbreak. A wide HBGA-binding spectrum is useful in understanding the epidemiology, host susceptibility and host range of GII.6 norovirus, supporting a need for continuous health attention and surveillance in different settings.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0950268820002538
Data availability statement
The datasets used and/or analysed during this study are available from the corresponding author on reasonable request.
Acknowledgements
The study was supported by the National Natural Science Foundation of China (grant numbers 31771007, 81773975); Natural Science Foundation of Guangdong Province, China (grant numbers 2019A1515010951); and Science and Technology Commission of Shanghai Municipality (grant numbers 201940428).
Conflicts of interest
None.
Ethics approval and consent to participate
Sample collection was approved by the Ethics Committee of Nanfang Hospital, Southern Medical University and informed consent was obtained from each involved individual.








