Introduction
Acanthocephalans (commonly named as spiny- or thorny-headed worms) are an important group of obligate endoparasites occurring in the alimentary track of all major vertebrate groups (Petrochenko and Skrjabin, Reference Petrochenko and Skrjabin1956; Yamaguti, Reference Yamaguti1963; Nickol, Reference Nickol1985; Naidu, Reference Naidu2012; Amin, Reference Amin2013), which are of veterinary, medical and economic importance in domestic animals, wildlife and humans (Petrochenko and Skrjabin, Reference Petrochenko and Skrjabin1956; Moore et al., Reference Moore, Fry and Englert1969; Nickol, Reference Nickol1985). Some acanthocephalan species of the genera Macracanthorhynchus Travassos, 1917, Moniliformis Travassos, 1915, Corynosoma Lühe, 1904, and Bolbosoma Porta, 1908, rarely Pseudoacanthocephalus Petrochenko, 1956 and Acanthocephalus Koelreuther, 1771, are recognized as zoonotic parasites associated with human acanthocephaliasis (Skrinnik et al., Reference Skrinnik, Likhotinskaya and Ocheret1958; Schmidt, Reference Schmidt1971; Leng et al., Reference Leng, Huang and Liang1983; Miyazaki, Reference Miyazaki1991; Berenji et al., Reference Berenji, Fata and Hosseininejad2007; Fujita et al., Reference Fujita, Waga, Kitaoka, Imagawa, Komatsu, Takanashi, Anbo, Anbo, Katuki, Ichihara, Fujimori, Yamasaki, Morishima, Sugiyama and Katahira2016).
The present knowledge regarding the basic molecular phylogenetic framework of the phylum Acanthocephala Rudolphi, 1808 remains far from complete. Previous studies indicated that the mitochondrial genomes play important roles in the phylogenetics, population genetics and species identification of acanthocephalans (Gazi et al., Reference Gazi, Kim, García-Varela, Park, Littlewood and Park2016; Song et al., Reference Song, Zhang, Deng, Ding, Liao and Liu2016; Pan and Jiang, Reference Pan and Jiang2018; Muhammad et al., Reference Muhammad, Suleman, Ma, Khan, Li, Zhao, Ahmad and Zhu2019a, Reference Muhammad, Suleman, Ma, Khan, Wu, Zhu and Li2019b, Reference Muhammad, Li, Suleman, Zhao, Bannai, Mohammad, Khan, Zhu and Ma2020a, Reference Muhammad, Suleman, Ahmad, Li, Zhao, Ullah, Zhu and Ma2020b, Reference Muhammad, Suleman, Khan, Li, Zhao, Ullah, Zhu and Ma2020c; Dai et al., Reference Dai, Yan, Li, Zhang, Liu, Gao, Ohiolei, Wu, Guo, Fu and Jia2022; Gao et al., Reference Gao, Yuan, Wu, Xiang, Xie, Song, Chen, Wu and Ou2022). However, the mitogenomes have been sequenced for only 29 acanthocephalan species, which belonged to 13 families in 6 orders. To date, there are approximately 98% of nominal species and 87% of genera in Acanthocephala with their mitogenomes unavailable so far, which somewhat hinder understanding the mitogenomic evolution and phylogenetics in Acanthocephala.
The order Polymorphida Petrochenko, 1956 currently includes 3 families Plagiorhynchidae Golvan, 1960, Centrorhynchidae Van Cleave, 1916 and Polymorphidae Meyer, 1931 (Amin, Reference Amin2013). Until now, the evolutionary relationships of the 3 families remain under debate. Some previous molecular analyses considered the Plagiorhynchidae and Polymorphidae to have an affinity (García-Varela et al., Reference García-Varela, de León, Aznar and Nadler2013; Gazi et al., Reference Gazi, Kim and Park2015; Muhammad et al., Reference Muhammad, Li, Suleman, Zhao, Bannai, Mohammad, Khan, Zhu and Ma2020a, Reference Muhammad, Suleman, Ahmad, Li, Zhao, Ullah, Zhu and Ma2020b, Reference Muhammad, Suleman, Khan, Li, Zhao, Ullah, Zhu and Ma2020c); however, other studies supported the Polymorphidae and Centrorhynchidae have closer relationship than the Plagiorhynchidae (Gazi et al., Reference Gazi, Kim, García-Varela, Park, Littlewood and Park2016; Song et al., Reference Song, Zhang, Gao, Cheng, Xie, Li and Wu2019; Muhammad et al., Reference Muhammad, Suleman, Ma, Khan, Li, Zhao, Ahmad and Zhu2019a, Reference Muhammad, Suleman, Ma, Khan, Wu, Zhu and Li2019b; Dai et al., Reference Dai, Yan, Li, Zhang, Liu, Gao, Ohiolei, Wu, Guo, Fu and Jia2022; Gao et al., Reference Gao, Yuan, Wu, Xiang, Xie, Song, Chen, Wu and Ou2022; Zhao et al., Reference Zhao, Yang, Lu, Ru, Wayland, Chen, Li and Li2023).
In Polymorphida, the Polymorphidae is the largest family and contains 12 genera, namely Andracantha Schmidt, Reference Schmidt1975, Ardeirhynchus Dimitrova and Georgiev, Reference Dimitrova and Georgiev1994, Arhythmorhynchus Lühe, 1911, Bolbosoma, Corynosoma, Diplospinifer Fukui, 1929, Filicollis Lühe, 1911, Ibirhynchus García-Varela, de León, Aznar et Nadler, 2011, Polymorphus Lühe, 1911, Profilicollis Meyer, 1931, Pseudocorynosoma Aznar, de León et Raga, 2006, and Southwellina Witenberg, 1932, with over 120 species reported from marine mammals, fish-eating marine birds and waterfowls worldwide (Deliamure, Reference Deliamure1968; Schmidt, Reference Schmidt1975; Dimitrova and Georgiev, Reference Dimitrova and Georgiev1994; Aznar et al., Reference Aznar, de León and Raga2006; García-Varela et al., Reference García-Varela, de León, Aznar and Nadler2011, Reference García-Varela, de León, Aznar and Nadler2013; Amin, Reference Amin2013). Several species of Bolbosoma and Corynosoma, for example, Bolbosoma cf. capitatum, Bolbosoma sp., Corynosoma villosum Van Cleave, 1953, C. strumosum (Rudolphi, 1802), C. validum Van Cleave, 1953 and Corynosoma sp. are recognized as parasites associated with human acanthocephaliasis (Beaver et al., Reference Beaver, Otsuji, Otsuji, Yoshimura, Uchikawa and Sato1983; Tada et al., Reference Tada, Otsuji, Kamiya, Mimori, Sakaguchi and Makizumi1983; Ishikura et al., Reference Ishikura, Takahashi, Sato, Kon, Oku, Kamiya, Ishikura, Yagi, Ishii, Yamamoto, Kamura, Kamiya and Kikuchi1996; Mori et al., Reference Mori, Maeba, Sekimata, Kobayashi, Suguri, Harada and Kagei1998; Hino et al., Reference Hino, Tsuchihashi, Kobayashi, Arizono and Kagei2002; Isoda et al., Reference Isoda, Kuroda, Shimizu and Okumura2006; Arizono et al., Reference Arizono, Kuramochi and Kagei2012; Fujita et al., Reference Fujita, Waga, Kitaoka, Imagawa, Komatsu, Takanashi, Anbo, Anbo, Katuki, Ichihara, Fujimori, Yamasaki, Morishima, Sugiyama and Katahira2016; Takahashi et al., Reference Takahashi, Ito, Sato, Goto, Kawamoto, Fujinaga, Yanagawa, Saito, Nakao, Hasegawa and Fujiya2016; Kaito et al., Reference Kaito, Sasaki, Goto, Matsusue, Koyama, Nakao and Hasegawa2019; Santoro et al., Reference Santoro, Palomba, Gili, Marcer, Marchiori and Mattiucci2021). Despite their significance, the mitochondrial genomes for representatives of Bolbosoma and Corynosoma have not been reported.
In the present study, the complete mitochondrial genomes of Bolbosoma nipponicum Yamaguti, 1939 and C. villosum (Acanthocephala: Polymorphidae) are sequenced and annotated for the first time, based on specimens collected from the northern fur seal Callorhinus ursinus (Linnaeus) (Mammalia: Carnivora) and the Pacific halibut Hippoglossus stenolepis (Schmidt) (Pleuronectiformes: Pleuronectidae) in Alaska, USA, which also represented the first mitogenome from the genera Bolbosoma and Corynosoma (Polymorphida: Echinorhynchidae). Additionally, in order to further clarify the evolutionary relationships of the 3 families Plagiorhynchidae, Centrorhynchidae and Polymorphidae in the order Polymorphida, phylogenetic analyses based on concatenated amino acid (AA) sequences of the 12 protein-coding genes (PCGs) of all available acanthocephalan mitogenomes were performed using Bayesian inference (BI) and maximum likelihood (ML), respectively.
Materials and methods
Species identification
The acanthocephalan specimens of B. nipponicum and C. villosum were collected from the intestine of subadult northern fur seals Callorhinus ursinus (Linnaeus) (Mammalia: Carnivora) and Hippoglossus stenolepis (Schmidt) (Pleuronectiformes: Pleuronectidae) in St. Paul Island, Alaska, USA, fixed and stored in 70% ethanol. The specimens were identified as B. nipponicum and C. villosum based on morphological features according to previous studies (Van Cleave, Reference Van Cleave1953; Margolis, Reference Margolis1956; Ru et al., Reference Ru, Yang, Chen, Kuzmina, Spraker and Li2022). Voucher specimens were deposited in the College of Life Sciences, Hebei Normal University, Hebei Province, China (B. nipponicum: HBNU–A-2022M002L; C. villosum: HBNU–A-2022F003L).
Molecular procedures
The total genomic DNA of each individual of B. nipponicum and C. villosum was extracted using the Magnetic Universal Genomic DNA Kit (DP705) (Tiangen Biotech, Beijing, China) according to the manufacturer's instructions: (1) cut the sample tissue into small pieces, add 300 μL tissue digestives GHA and 20 μL Proteinase K, and grind the tissue thoroughly; (2) transfer the above-treated sample solution of 300 μL into a new 1.5 mL centrifuge tube; (3) add 300 μL lysate GHL and 300 μL isopropyl alcohol, shake and mix well; (4) add 15 μL magnetic bead suspension GH, shake and mix for 1 min, stand for 9 min in total, shake and mix for 1 min each 3 mins; (5) the centrifuge tube was placed on the magnetic rack and stood for 30 s. After the magnetic bead was completely absorbed, the liquid was carefully absorbed, and the DNA solution was transferred to the new centrifuge tube. The genomic DNA sample was fragmented by sonication to a size of 350 bp in preparation for genomic library.
A total of 20 GB of gene library data for each sample of B. nipponicum and C. villosum were yielded using the Pair-End 150 sequencing method on the Illumina NovaSeq 6000 platform by Novogene (Tianjin, China). Program GetOrganelle v1.7.2a (Jin et al., Reference Jin, Yu, Yang, Song, dePamphilis, Yi and Li2020) was employed to reconstruct the mitochondrial genomes of these acanthocephalans. The locations of PCGs, transfer RNA (tRNA), and ribosomal RNA (rRNA) in the generated mitochondrial genomes were roughly annotated using MitoZ v2.4 (Meng et al., Reference Meng, Li, Yang and Liu2019) and web-servers MITOS (http://mitos.bioinf.uni-leipzig.de/index.py). All PCGs were confirmed manually using the ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/) based on the invertebrate mitochondrial genetic code. Transfer RNA genes were additionally identified using BLAST-based on a database of the existing tRNA sequences of Acanthocephala. The secondary structures of tRNAs were predicted by the ViennaRNA module (Gruber et al., Reference Gruber, Bernhart and Lorenz2015) and then manually corrected building on MitoS2 (Bernt et al., Reference Bernt, Merkle, Ramsch, Fritzsch, Perseke, Bernhard, Schlegel, Stadler and Middendorf2007) and RNAstructure v6.3 (Reuter and Mathews, Reference Reuter and Mathews2010). The CGView online server V1.0 (http://stothard.afns.ualberta.ca/cgview_server/) was used to generate the circular genomic maps. The base composition, amino acid usage and relative synonymous codon usage (RSCU) were calculated by Python script (details see the Supplementary material), which refers to codon adaptation index (Cock et al., Reference Cock, Antao, Chang, Chapman, Cox, Dalke, Friedberg, Hamelryck, Kauff, Wilczynski and de Hoon2009). Strand asymmetry was calculated using the formulae: AT-skew = (A − T)/(A + T); GC-skew = (G − C)/(G + C). The complete mitochondrial genomes of B. nipponicum and C. villosum obtained herein were deposited in the GenBank database (http://www.ncbi.nlm.nih.gov, under the accession numbers Corynosoma villosum: OR468095, Bolbosoma nipponicum: OR468096).
Phylogenetic analyses
Phylogenetic analyses were conducted based on concatenated amino acid (AA) sequences of the 12 PCGs using BI and ML, respectively. Two species of Bdelloidea, Rotaria rotatoria (NC013568.1) and Philodina citrina (FR856884.1) were chosen as the out-group. The in-group included the newly sequenced B. nipponicum and C. villosum and the other 29 species of acanthocephalans with mitogenomic data. Detailed information on representatives included in the present phylogeny was provided in Table 1. For phylogenetic analyses, PhyloSuite was used to collect all mitogenomic sequences from GenBank files, standardize annotation and extract mitogenomic data (Zhang et al., Reference Zhang, Gao, Jakovlić, Zou, Zhang, Li and Wang2020). The extracted amino acid sequences of all 12 PCGs were aligned in MAFFT v7.313 under iterative refinement method of E-INS-I (Katoh and Standley, Reference Katoh and Standley2013). The AAs dataset was concatenated into a single alignment by PhyloSuite, respectively. Substitution models were compared and selected according to the Bayesian Information Criterion (BIC) by using ModelFinder (Kalyaanamoorthy et al., Reference Kalyaanamoorthy, Minh, Wong, von Haeseler and Jermiin2017). The VT + F + I + G4 was identified as the optimal amino acid substitution model for both partitions (partition1: cox1, cox2, nad1; partition2: all other genes). Bayesian Information Criterion analysis settings were lser nst = 6, rates = invgamma, mcmc ngen = 5 000 000, printfreq = 1000, samplefreg = 100, nchains = 4, nruns = 2. The analysis continued until the average standard deviation of split frequencies was lower than 0.01. The first 25% of trees were treated as ‘burn-in’. For ML analysis, 1000 bootstrap replicates were used to calculate the bootstrap of the program in IQTREE v2.1.2, keep the default values for other parameters (Golombek et al., Reference Golombek, Tobergte and Struck2015; Minh et al., Reference Minh, Schmidt, Chernomor, Schrempf, Woodhams, von Haeseler and Lanfear2020). The iTOL v6.1.1 was used to visualize the phylogeny and architecture using files generated by PhyloSuite (Letunic and Bork, Reference Letunic and Bork2021).
Table 1. Detailed information of representatives with their mitogenomic data included in the present phylogeny

Bolbosoma nipponicum and Corynosoma villosum in the present study are indicated in bold.
Results
General characterization of the complete mitogenomes of B. nipponicum and C. villosum
Gene content and organization of mitogenomes
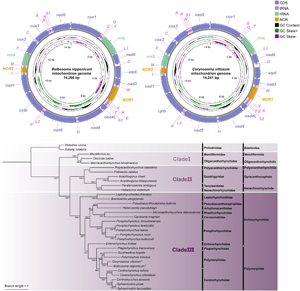
The complete mitochondrial genomes of B. nipponicum and C. villosum are 14 296 and 14 241 bp in length, respectively, and both contain 36 genes, including 12 PCGs (missing atp8) (cox1–3, cytb, nad1–6, nad4L and atp6), 22 tRNA genes and 2 rRNA genes (rrnL and rrnS) (Fig. 1, Table 2). All genes are transcribed from the same strand. Two non-coding regions (NCR1 and NCR2), are present in the same positions in the mitogenomes of B. nipponicum and C. villosum (NCR1 is 776 bp in B. nipponicum vs 676 bp in C. villosum, both located between trnW and trnV; NCR2 is 301 bp in B. nipponicum vs 318 bp in C. villosum, both between trnI and trnM) (Fig. 1; Table 2). The overall A + T contents in the mitogenomes of B. nipponicum and C. villosum are 60.88% and 60.99%, respectively (Table 3), both displaying strong A + T bias.

Figure 1. Gene maps of the mitochondrial genomes of Bolbosoma nipponicum and Corynosoma villosum. All genes are transcribed in the clockwise direction on the same strand, and 22 tRNA genes are designated by the 1-letter code with numbers differentiating each of the 2 tRNAs serine and leucine. The outermost circle shows the GC content and the innermost circle shows the GC skew.
Table 2. Organization of the mitochondrial genomes of Bolbosoma nipponicum and Corynosoma villosum

Ini/Ter cod: initial/terminal codons, Int seq: intergenic sequences.
Bolbo.
Table 3. Base composition and skewness in the mitogenomes of Bolbosoma nipponicum and Corynosoma villosum

Bolbosoma nipponicum and Corynosoma villosum in the present study are indicated in bold.
Protein-coding genes and codon usage
The 12 PCGs of the mitogenomes of B. nipponicum and C. villosum are 10 468 bp and 10 389 bp in total length (excluding termination codons), and ranged from 270 bp (nad4L) to 1,647 bp (nad5) in size (Table 2). Among the 12 PCGs of B. nipponicum, 8 genes (cox2, cox3, cytb, nad2, nad3, nad4L, nad5 and nad6) used GTG as the start codon, whereas 2 genes (nad1 and nad4) used ATG. ATT and TTG were used by the atp6 and cox1 genes, respectively. TAG was the most commonly used termination codon for 6 genes (atp6, cox1, nad1, nad2, nad5 and nad6); 4 genes (cox2, cox3, cytb and nad4L) used TAA. The incomplete termination codon T was inferred for the nad3 and nad4 genes (Table 2).
Among the 12 PCGs of C. villosum, 9 genes (cox1, cox2, cox3, cytb, nad1, nad2, nad4L, nad5 and nad6) used GTG as the start codon, whereas 3 genes (atp6, nad3 and nad4) used ATG. TAA was the most commonly used termination codon for 6 genes (atp6, cox1, cox2, cox3, nad2 and nad4L); 3 genes (cytb, nad1 and nad5) used TAG. The incomplete termination codon T was inferred for the nad3, nad4 and nad6 genes (Table 2). RSCU is summarized in Fig. 2.

Figure 2. Relative synonymous codon usage (RSCU) of Bolbosoma nipponicum and Corynosoma villosum. Codon families (in alphabetical order) are provided below the horizontal axis. Values on the top of each bar represent amino acid usage in percentage.
Transfer and ribosomal RNAs
In the mitogenomes of B. nipponicum and C. villosum, 22 tRNAs are identified with lengths ranging from 50 to 61 bp in B. nipponicum, and from 52 to 64 bp in C. villosum (Fig. 1, Table 2). Their anticodon secondary structures are provided (Figs 3, 4). In the 22 tRNAs of B. nipponicum and C. villosum, 5 tRNAs (trnR, trnN, trnF, trnW and trnV) were predicted to be folded into typical cloverleaf secondary structure, 2 tRNAs (trnQ and trnS1) lacked dihydorourdine (DHU) arm, the remaining 15 tRNAs lacked TΨC arm (Figs 3, 4, Table 2).

Figure 3. The predicted secondary structures of 22 tRNAs in the mitogenome of Bolbosoma nipponicum (Watson–Crick bonds indicated by lines, GU bonds indicated by dots, red bases representing anticodons). The tRNAs are labelled with the abbreviations of their corresponding amino acids according to the IUPAC-IUB code.

Figure 4. The predicted secondary structures of 22 tRNAs in the mitogenome of Corynosoma villosum (Watson–Crick bonds indicated by lines, GU bonds indicated by dots, red bases representing anticodons). The tRNAs are labelled with the abbreviations of their corresponding amino acids according to the IUPAC-IUB code.
Two rRNAs (rrnL and rrnS) were identified in the mitogenomes of B. nipponicum and C. villosum (rrnL is 913 bp in B. nipponicum vs 914 bp in C. villosum, both located between trnY and trnL1; rrnS is 585 bp in B. nipponicum vs 584 bp in C. villosum, both located between trnM and trnF) (Fig. 1, Table 2).
Gene order
The arrangements of the 36 genes in the mitogenomes of B. nipponicum and C. villosum are identical, both in the following order: cox1, trnG, trnQ, trnY, rrnL, trnL1, nad6, trnD, atp6, nad3, trnW, trnV, trnK, trnE, trnT, trnS2, nad4L, nad4, trnH, nad5, trnL2, trnP, cytb, nad1, trnI, trnM, rrnS, trnF, cox2, trnC, cox3, trnA, trnR, trnN, trnS1, nad2 (Fig. 5).

Figure 5. Comparison of the linearized mitochondrial genome arrangement for acanthocephalans species. All genes are transcribed in the same direction from left to right. The tRNAs are labelled by single-letter code for the corresponding amino acid. Bolbosoma nipponicum and Corynosoma villosum are indicated using asterisk (*).
Phylogenetic analyses
In the present study, phylogenetic trees based on concatenated amino acid sequences of the 12 PCGs using ML and BI methods have nearly same topologies, except the supported value of some branches, which all showed that the representatives of the phylum Acanthocephala were divided into 3 large monophyletic clades (Figs 6). Clade I includes Moniliformis sp., Macracanthorhynchus hirudinaceus and Oncicola luehei, which represents the class Archiacanthocephala at the most basal position of the phylogenetic trees. Clade II contains the representatives of the class Eoacanthocephala (Acanthogyrus bilaspurensis, A. cheni, Hebesoma violentum, Paratenuisentis ambiguus and Pallisentis celatus) and Polyacanthorhynchus caballeroi (belonging to the class Polyacanthocephala). Clade III is composed of the representatives of the class Palaeacanthocephala, of which the order Echinorhynchida (including Cavisoma magnum, Echinorhynchus truttae, Pseudoacanthocephalus bufonis, Brentisentis yangtzensis, Pomphorhynchus spp., Leptorhynchoides thecatus and Micracanthorhynchina dakusuiensis) is a non-monophyletic group, but the order Polymorphida (including Plagiorhynchus transversus, Polymorphus minutus, Southwellina hispida, Centrorhynchus spp., Sphaerirostris spp., Bolbosoma nipponicum and Corynosoma villosum) is a monophyletic group. In the order Polymorphida, the family Polymorphidae is a sister to the family Centrorhynchidae. Bolbosoma nipponicum and Corynosoma villosum clustered together with strong nodal support (BPP = 1, ML-BP = 100) in all BI and ML trees.

Figure 6. Phylogenetic analyses of Acanthocephala inferred from ML and BI methods based on concatenated amino acid sequences of 12 PCGs of mitochondrial genomes. Rotaria rotatoria and Philodina citrina were chosen as the out-group. Bootstrap values ⩾70 and Bayesian posterior probabilities values ⩾0.70 are shown in the phylogenetic trees. Bolbosoma nipponicum and Corynosoma villosum are indicated using asterisk (*).
Discussion
The order Polymorphida is a large group of Acanthocephala, containing over 370 nominal species mainly parasitic in birds and mammals, rarely in reptiles (Amin, Reference Amin2013; Zhao et al., Reference Zhao, Muhammad, Chen, Ma, Suleman and Li2020; Ru et al., Reference Ru, Yang, Chen, Kuzmina, Spraker and Li2022). According to the traditional classification, Polymorphida was divided into 3 families (Centrorhynchidae, Polymorphidae, and Plagiorhynchidae), including 24 genera (Amin, Reference Amin2013). However, only 8 species representing 5 genera with their mitochondrial genomes have been documented (Gazi et al., Reference Gazi, Kim and Park2015, Reference Gazi, Kim, García-Varela, Park, Littlewood and Park2016; Muhammad et al., Reference Muhammad, Suleman, Ma, Khan, Li, Zhao, Ahmad and Zhu2019a, Reference Muhammad, Suleman, Ma, Khan, Wu, Zhu and Li2019b, Reference Muhammad, Li, Suleman, Zhao, Bannai, Mohammad, Khan, Zhu and Ma2020a, Reference Muhammad, Suleman, Ahmad, Li, Zhao, Ullah, Zhu and Ma2020b; Sarwar et al., Reference Sarwar, Zhao, Kibet, Sitko and Nie2021). In Polymorphida, the size of mitochondrial genomes of B. nipponicum and C. villosum is similar to that of the polymorphid species Polymorphus minutus and Southwellina hispida; however, the overall A + T contents in the mitogenomes of B. nipponicum and C. villosum are slightly lower than that of P. minutus and S. hispida (60.88–60.99% in B. nipponicum and C. villosum vs 63.9–64.4% in the latter 2 species) (Gazi et al., Reference Gazi, Kim and Park2015; Sarwar et al., Reference Sarwar, Zhao, Kibet, Sitko and Nie2021), but distinctly higher than that of centrorhynchid species (54.5–58.1%) (Muhammad et al., Reference Muhammad, Suleman, Ma, Khan, Li, Zhao, Ahmad and Zhu2019a, Reference Muhammad, Suleman, Ma, Khan, Wu, Zhu and Li2019b, Reference Muhammad, Li, Suleman, Zhao, Bannai, Mohammad, Khan, Zhu and Ma2020a, Reference Muhammad, Suleman, Ahmad, Li, Zhao, Ullah, Zhu and Ma2020b; Gazi et al., Reference Gazi, Kim, García-Varela, Park, Littlewood and Park2016).
Previous studies indicated that the arrangements of the 12 PCGs and 2 rRNAs in the phylum Acanthocephala are highly conserved, while the position of tRNAs usually varied among different families, genera or species (Gazi et al., Reference Gazi, Sultana, Min, Park, García-Varela, Nadler and Park2012, Reference Gazi, Kim, García-Varela, Park, Littlewood and Park2016; Dai et al., Reference Dai, Yan, Li, Zhang, Liu, Gao, Ohiolei, Wu, Guo, Fu and Jia2022; Gao et al., Reference Gao, Yuan, Wu, Xiang, Xie, Song, Chen, Wu and Ou2022; Zhao et al., Reference Zhao, Yang, Lu, Ru, Wayland, Chen, Li and Li2023). In the family Polymorphidae, the arrangements of the 36 genes in the mitogenomes of B. nipponicum and C. villosum are identical to that of P. minutus (Sarwar et al., Reference Sarwar, Zhao, Kibet, Sitko and Nie2021), but the positions of some tRNAs are different from that of the polymorphid species S. hispida (Gazi et al., Reference Gazi, Kim and Park2015) and all of the species of Centrorhynchidae and Plagiorhynchidae (Muhammad et al., Reference Muhammad, Suleman, Ma, Khan, Li, Zhao, Ahmad and Zhu2019a, Reference Muhammad, Suleman, Ma, Khan, Wu, Zhu and Li2019b, Reference Muhammad, Li, Suleman, Zhao, Bannai, Mohammad, Khan, Zhu and Ma2020a, Reference Muhammad, Suleman, Ahmad, Li, Zhao, Ullah, Zhu and Ma2020b; Gazi et al., Reference Gazi, Kim, García-Varela, Park, Littlewood and Park2016).
Recent mitogenomic phylogenies brought substantial changes to the traditional classification of Acanthocepha. However, phylogenetic relationships within many lineages of the class Palaeacanthocephala remain insufficiently resolved, due to large numbers of taxa (i.e. the order Heteramorphida, and the families of llliosentidae, Isthmosacanthidae, Heteracanthocephalidae, Fessisentidae, Diplosentidae, Transvenidae, Spinulacorpidae Hypoechinorhynchidae and Gymnorhadinorhynchidae of the order Echinorhynchida) that have not been included yet. Although all of the 3 family-level taxa (Plagiorhynchidae, Centrorhynchidae and Polymorphidae) of the order Polymorphida have been included in some previous mitogenomic phylogenetic studies, very limited genus/species-level taxa have been covered in each family. The evolutionary relationships of the 3 families in Polymorphida and its included genera of each family remain unsolved.
The present mitogenomic phylogenies showed that the order Polymorphida is a monophyletic group, but rejected the monophyly of the order Echinorhynchida, in agreement with the previous studies (García-Varela and Nadler, Reference García-Varela and Nadler2006; García-Varela and de León, Reference García-Varela and de León2008; Verweyen et al., Reference Verweyen, Klimpel and Palm2011; García-Varela et al., Reference García-Varela, de León, Aznar and Nadler2013; Braicovich et al., Reference Braicovich, Lanfranchi, Farber, Marvaldi, Luque and Timi2014). The previous molecular phylogenetic results using single or several concatenated genetic markers (i.e. 18S, 18S + 28S + cox1) (Near et al., Reference Near, Garey and Nadler1998; García-Varela et al., Reference García-Varela, de León, Aznar and Nadler2013) and some recent mitogenomic phylogenies (Muhammad et al., Reference Muhammad, Li, Suleman, Zhao, Bannai, Mohammad, Khan, Zhu and Ma2020a, Reference Muhammad, Suleman, Ahmad, Li, Zhao, Ullah, Zhu and Ma2020b, Reference Muhammad, Suleman, Khan, Li, Zhao, Ullah, Zhu and Ma2020c; Sarwar et al., Reference Sarwar, Zhao, Kibet, Sitko and Nie2021) indicated that the Plagiorhynchidae is a sister to the Polymorphidae or Centrorhynchidae. However, the present phylogenetic results strongly displayed that the Polymorphidae and Centrorhynchidae are more closely related to each other than to the Plagiorhynchidae (Gazi et al., Reference Gazi, Kim, García-Varela, Park, Littlewood and Park2016; Muhammad et al., Reference Muhammad, Suleman, Ma, Khan, Li, Zhao, Ahmad and Zhu2019a, Reference Muhammad, Suleman, Ma, Khan, Wu, Zhu and Li2019b; Song et al., Reference Song, Zhang, Gao, Cheng, Xie, Li and Wu2019; Dai et al., Reference Dai, Yan, Li, Zhang, Liu, Gao, Ohiolei, Wu, Guo, Fu and Jia2022; Gao et al., Reference Gao, Yuan, Wu, Xiang, Xie, Song, Chen, Wu and Ou2022; Zhao et al., Reference Zhao, Yang, Lu, Ru, Wayland, Chen, Li and Li2023). Although Zhao et al.’ (Reference Zhao, Yang, Lu, Ru, Wayland, Chen, Li and Li2023) study also suggests a close affinity between the Polymorphidae and Centrorhynchidae, there are only 2 representatives of the Polymorphidae in their phylogeny, and the supported value for the close phylogenetic relationship of the Polymorphidae and Centrorhynchidae is weak. However, the present phylogenetic study including 2 additional genus-level taxa of the Polymorphidae, showed strong support for the close affinity between the Polymorphidae and Centrorhynchidae in both ML (BS = 98) and BI (BPP = 1).
Our phylogenetic results also revealed the genus Bolbosoma has a sister relationship with Corynosoma based on the mitogenomic data for the first time, in concordance with previous studies based on single or several concatenated genetic markers (García-Varela et al., Reference García-Varela, de León, Aznar and Nadler2013; Presswell et al., Reference Presswell, Garcia-Varela and Smales2018; Ru et al., Reference Ru, Yang, Chen, Kuzmina, Spraker and Li2022). Moreover, the present findings further clarified the phylogenetic relationships of the 3 families Plagiorhynchidae, Centrorhynchidae and Polymorphidae, enriched the mitogenome data of the phylum Acanthocephala (especially the order Polymorphida), and also provided the resource of genetic data for diagnosing these 2 pathogenic parasites of human acanthocephaliasis.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182023001099.
Data availability
The complete mitochondrial genomes of B. nipponicum and C. villosum obtained herein were deposited in the GenBank database (http://www.ncbi.nlm.nih.gov, under the accession numbers Corynosoma villosum: OR468095, Bolbosoma nipponicum: OR468096). Voucher specimens of B. nipponicum and C. villosum were deposited in the College of Life Sciences, Hebei Normal University, Hebei Province, China.
Acknowledgements
The authors are grateful to Professor Hideo Hasegawa (Faculty of Medicine, Oita University, Japan) for providing important literature. The authors wish to thank the people of the Aleut community and the National Marine Mammal Laboratory scientists for providing helps during collection of the parasites.
Author's contributions
D. X. L. and L. L. contributed to the study design and conducted the phylogenetic analyses. D. X. L., R. J. Y., H. X. C. and L. L. sequenced and analysed genetic data. H. X. C. and L. L. identified the acanthocephalan specimens. T. A. K. and T. R. S. provided acanthocephalan specimens. D. X. L. and L. L. wrote the manuscript. All authors read and approved the final manuscript.
Financial support
This study was supported by the National Natural Science Foundation of China (Grant No. 31872197) and the National Key R&D Program of China (Grant No. 2022YFC2601200).
Competing interest
None.
Ethical standards
This study was conducted under the protocol of Hebei Normal University. All applicable national and international guidelines for the protection and use of animals were followed.