Rashes are extremely common in newborns and can be a significant source of parental concern. Although most rashes are transient and benign [Reference O‘Connor, McLaughlin and Ham1], some may be indicative of an underlying disorder such as bacterial or viral infection. Neonatal exanthema is a self-limiting rash characterized by small sterile erythematous papules or vesicles, and sometimes pustules on the trunk, extremities and the face. The aetiology of the condition is uncertain but an infectious origin has been shown for a minority of patients and an association with Listeria spp., group B Streptococcus and Staphylococcus aureus. [Reference Casacci2] Gram-negative bacterial infection is rare and a maculopapular appearance of rash is uncommon, although Klebsiella pneumoniae has been implicated in a case of a neonate with an exanthematous rash apparently acquired from the mother [Reference Casacci2]. There are no reports of outbreaks due to this organism associated with rash. In this report we describe an outbreak of neonatal sepsis caused by K. pneumoniae in a level III neonatal intensive care unit (NICU) of a developing country, where the majority of affected babies presented with a maculopapular rash.
In July 2008, an extended-spectrum β-lactamase (ESBL) producing K. pneumoniae was isolated from the blood of a term, 2-day-old inborn neonate, admitted to the NICU with respiratory distress. The baby developed a maculopapular rash soon after admission. An outbreak was declared when six cases were identified within 5 days. The case definition was (i) admission to the NICU, (ii) presence of two or more of the following: respiratory symptoms, thermal instability, poor feeding, lethargy, abdominal distension, presence of maculopapular rash, (iii) blood/CSF culture positive for ESBL K. pneumoniae.
Blood culture was performed with a Bactec 9050 system (Becton Dickinson, USA). The identification of isolates was confirmed using the ID32E kit (bioMérieux, France). Antibiotic susceptibility was determined by the Kirby–Bauer disc diffusion method and interpreted according to Clinical Laboratory Standards Institute (CLSI) Guidelines [3]. ESBL production was confirmed by the CLSI phenotypic confirmatory test [3]. Environmental sampling of 23 sites in the labour room complex and NICU was undertaken on day 10 of the outbreak.
The DNA banding patterns of 12 K. pneumoniae isolates (Fig. 1), resolved by pulsed-field gel electrophoresis (PFGE) following the Pulse Net protocol (CDC, USA) [4] were interpreted according to Tenover et al. [Reference Tenover, Arbeit and Goering5].

Fig. 1. PFGE of XbaI-digested genomic DNA of the Klebsiella pneumoniae isolates. Marker (M): Salmonella serotype Braenderup H9812 as reference standard. Lanes 1a, 1b, 2a, 2b, 3–7, predominant outbreak strain. Lane 8, different. Lanes 9, 10, distinct from the predominant outbreak strain.
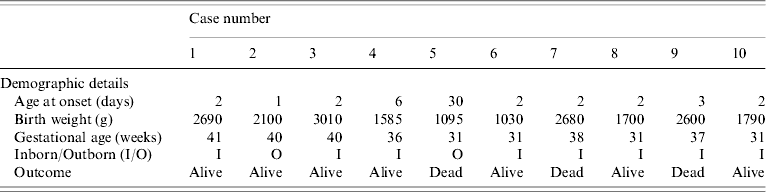
The outbreak lasted for 2 weeks and ten babies developed septicaemia with K. pneumoniae. Table 1 shows that there were eight cases of early onset sepsis, of which seven (cases 1, 4, 6–10) were inborn babies with vaginal delivery. Cases 1–7 developed an exanthematous rash which was maculopapular, <1 cm in size, circumscribed and non-blanching on application of pressure. It appeared first on the face and trunk and spread to the extremities except the palms of hands and soles of feet. The rash was not present at birth and subsided 2–3 days after starting antibiotics. At no stage did it evolve into a vesicle or pustule. During the course of the adoption of control measures, three further cases (cases 8–10) were documented but without rash.
Table 1. Demographic details of neonates with Klebsiella pneumoniae infections
Thirteen isolates of K. pneumoniae were obtained from ten babies: 11 were from blood culture (two cultures from the index case), one from endotracheal aspirate and one from pus. All the isolates were ESBL producers. The antibiograms were uniform except that, in contrast to the first seven cases, the later isolates were resistant to amikacin. All environmental surveillance cultures were negative for K. pneumoniae.
Molecular typing showed that nine isolates from the first seven cases were identical. This was identified as the predominant outbreak strain. The blood isolate from case 8 could not be typed owing to autodegradation of the DNA. The endotracheal isolate from this patient was distinct from all other isolates while the strains from cases 9 and 10 were indistinguishable from each other.
This was the first such outbreak in our unit in 8 years. The majority (8/10) of cases were very young babies (aged <3 days). The unusual feature we observed was the occurrence of maculopapular rash in the first seven cases infected with the predominant outbreak strain of K. pneumoniae. This finding was so distinctive that case 7 was identified as part of the outbreak, even before the blood culture report was available. The cause of the rash is unclear. Careful evaluation of maternal and perinatal history revealed that there was no history of skin disease, or significant systemic disease which could account for the rash. Maternal serology was negative for TORCH infections. None of the affected babies had eosinophilia which has been found in some cases of erythema neonatorum [Reference Tanioka, Honda and Takagaki6]. The selective excretion of anti-inflammatory cytokine interleukin 10 has been cited as a possible cause supporting the view that the erythema is an inflammatory skin reaction to microbial colonization at birth [Reference Takahashi7].
Various environmental sources have been implicated in hospital outbreaks of K. pneumoniae but often a primary source cannot be identified [Reference Gastmeier8]. In the present study, all environmental sources sampled did not yield K. pneumoniae. Nevertheless, sanitation and infection control protocols were reinforced. As the NICU serves a large patient population and is the only level III unit in the entire state, closure of the unit was not possible. The presence of a microbiologist within the unit itself was a key factor in the detection of the outbreak. While neonatal rash is caused by a wide variety of causes, both infectious and non-infectious, including drug allergy and insect bites, the standard literature [Reference Lucky, Eichenfield, Freiden and Esterly9, Reference Freiden, Howard, Eichenfield, Freiden and Esterly10] reveals that most of the rashes range from vesicle and pustule to bullae and erosions. One possible differential diagnosis is erythema toxicum neonatorum which may present as erythematous macules but this was excluded based on the clinical presentation. A limitation of our study was that scraping from the skin lesion was not studied for the presence of cellular infiltrates or microorganisms. Given the isolation of the same strain of K. pneumoniae from all the affected babies, an infectious aetiology for the rash appears likely. Further studies such as analysis of extracellular products including proteases and epidermolytic toxins may provide more information. In conclusion, the present study highlights that neonatal sepsis due to K. pneumoniae may present with a maculopapular rash. A unit in a developing country may not have ready access to molecular analysis but the involvement of different strains of the same organism in the time span of an outbreak may be distinguished by paying close attention to clinical presentation.
ACKNOWLEDGEMENTS
The authors thank Mr Deepak Singh for technical help.
DECLARATION OF INTEREST
None.




