Introduction
Salmonella is one of the noteworthy foodborne pathogens worldwide. Salmonella is ubiquitous with wide host range due to its capacity to survive in adverse environments. Most of the studies on public health implications of Salmonella in wildlife have an emphasis on amphibians and reptiles [Reference Stirling1, Reference Erdozain2]. The role of captive and free-range wildlife mammals and associated caretakers in the epidemiology of Salmonella is a domain that has rarely been investigated. Various serovars of public health significance including serovar Typhimurium and Newport and antimicrobial resistant strains have been reported from turtle, deer, wild birds and water samples [Reference Gorski3–Reference Kang5]. Clonally related isolates of different serovars of public health significance in captive wild mammals and associated environments were recently reported in the USA [Reference Farias6]. Very limited studies have reported the same occurrence of Salmonella serovars from humans and wildlife species, supporting that wild animals serve as reservoirs for salmonellosis in humans [Reference Kapperud7, Reference Handeland8]. Another serious concern in Salmonella is the growing antimicrobial resistance and the dissemination of multidrug-resistant (MDR) strains. Enterobacterial repetitive intergenic consensus (ERIC) fingerprinting is effective than pulsed-field gel electrophoresis (PFGE) and it is useful for subtyping Salmonella serovars, where similar PFGE patterns occur [Reference Fendri9]. ERIC-PCR was found effective over many other molecular typing methods [Reference Lim10, Reference Kumar11]. Besides, ERIC-PCR is a simple and cost-effective technique than PFGE. The aim of this study was to determine the occurrence, serovar distribution, antimicrobial susceptibility patterns and genotypic relatedness of Salmonella isolates recovered from faecal samples of captive wildlife, their caretakers, feed and water in India.
Materials and methods
The study was carried out in four zoological gardens and wildlife enclosures, viz., Nainital Zoo, Nainital, Uttarakhand; Kanpur Zoo, Kanpur, Uttar Pradesh; Deer Park, Indian Veterinary Research Institute, Izatnagar, Bareilly, Uttar Pradesh; Post Graduate Research Institute in Animal Sciences, Chennai, Tamilnadu, India. A total of 370 samples comprising 314 fresh faecal samples of apparently healthy captive animals (40 species) (Tables 1–3), 30 stool swabs from animal caretakers and 26 feed and water samples were collected. Briefly, 10 g samples were pre-enriched in 2% buffered peptone water (BPW) (HiMedia, India), at 37 °C for 16–18 h. The broth culture (100 µl) was transferred to 10 ml Tetrathionate Broth (HiMedia) and incubated at 37 °C for 24 h. A loopful of the suspension was streaked onto Hektoen Enteric Agar (HEA) (HiMedia) and incubated at 37 °C for 24 h [Reference Babu12]. Selected presumptive Salmonella colonies were inoculated onto triple sugar iron (TSI) agar (HiMedia) and Christensen's urea slants (HiMedia) and incubated at 37 °C for 24 h. All presumptive Salmonella isolates were submitted to the National Salmonella Centre (ICAR-IVRI, Bareilly, India) for serotyping. Genomic DNA was isolated directly from 314 faecal samples of captive wildlife by QIAamp DNA Stool Mini Kit (Qiagen, Germany). The extracted DNA was subjected to PCR as described by Lin and Tsen, [Reference Lin and Tsen13]. Pearson's chi-square and Fisher exact test were employed to compare the prevalence of Salmonella spp. among different zoos and various sample groups viz., wild ruminants, wild non-ruminants, wild birds, caretakers, feed and water (SPSS 22.0 version). The differences among various zoos and sample groups were considered significant at P < 0.05. The antimicrobial resistance profiles of Salmonella enterica isolates were tested to a panel of 23 different antibiotics (BD Diagnostics, Sparks, MD, USA) belonging to 10 classes using the Kirby–Bauer disc diffusion method. The antibiotic used were aminoglycosides-streptomycin (S, 10 µg), gentamicin (Gm, 10 µg), kanamycin (K, 30 µg), amikacin (Ak, 30 µg); colistin (Cl, 10 µg); cephalosporins- cefotaxime (Ctx, 30 µg), ceftazidime (Caz, 30 µg), ceftazidime + clavulanic acid (30/10 µg) and cefotaxime + clavulanic acid (30/10 µg); macrolides-erythromycin (E, 15 µg); fluoroquinolones-enrofloxacin (Ex, 10 µg), ciprofloxacin (Cip, 5 µg), ofloxacin (Of, 5 µg); monobactam-aztreonam (Atm, 30 µg); carbapenem antibiotics-imipenem (Ipm, 10 µg), meropenem (Mem, 10 µg), ertapenem (Etp, 10 µg); penicillins-carbenicillin (Cb, 100 µg), amoxicillin with clavulanic acid (Amc, 10 µg), ampicillin (Am, 10 µg); tetracycline (Te, 30 µg); sulphonamides (sulphamethoxazole with trimethoprim (Sxt, 10 µg)) and others-nitrofurantoin (F/M, 100 µg). Isolates showing resistance to three or more classes of antimicrobials were classified as MDR. Multiple antibiotic resistance index (MARI) for each resistance pattern was calculated.
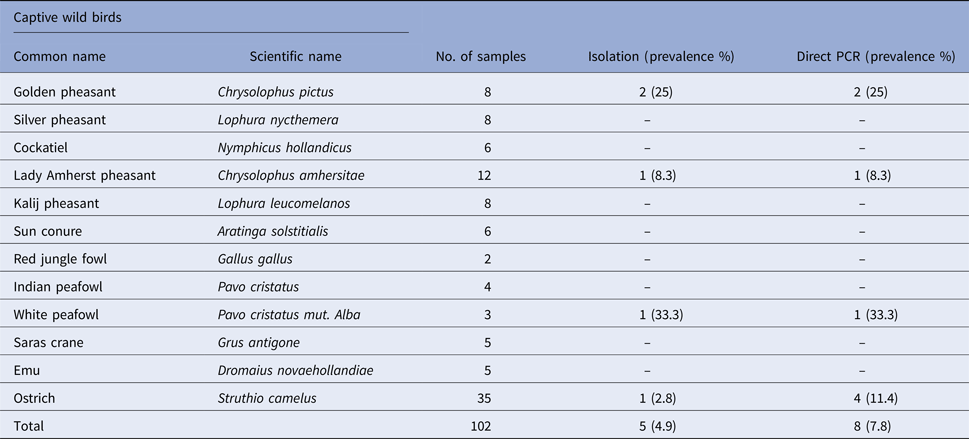
Table 1. Prevalence of Salmonella spp. in faecal samples collected from captive wild ruminants
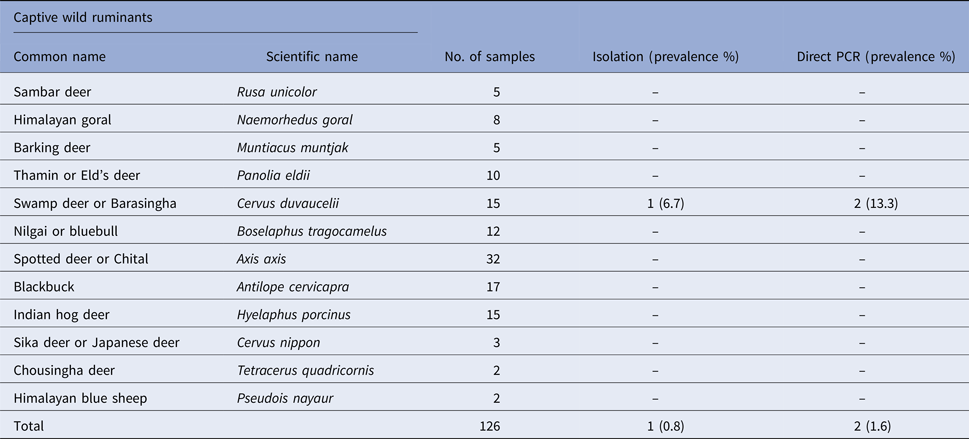
Table 2. Prevalence of Salmonella spp. in faecal samples collected from captive wild non-ruminants
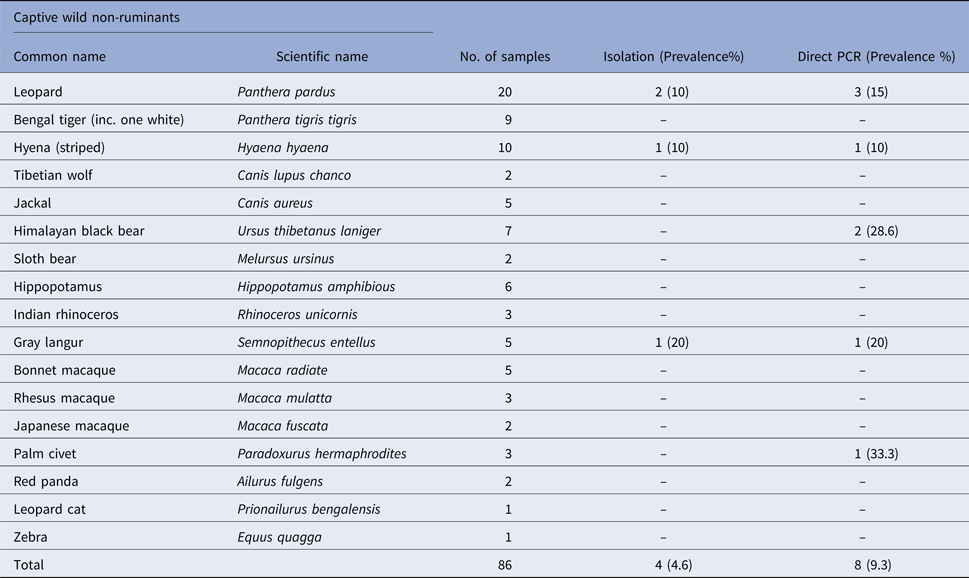
Table 3. Prevalence of Salmonella spp. in faecal samples collected from captive wild birds
The ERIC-PCR assay was performed as per Campioni et al. [Reference Campioni14]. The oligonucleotide primers described in a previous study was used [Reference Versalovic15]. The PCR reaction mixture for amplification consisted of 12.5 µl of 2× PCR master mixtures (ThermoFisher Scientific), 1 µl (10 pmol/μl) of each primer (Eurofins, India), 2 µl of DNA template and nuclease-free water to make final volume up to 25 µl. The program used for the ERIC-PCR are as follows: initial denaturation at 94 °C for 7 min followed by 30 cycles each of denaturation at 94 °C for 30 s, annealing at 52 °C for 1 min and extension at 65 °C for 8 min followed by final extension at 65 °C for 10 min. The PCR was performed in a Thermal Cycler (Eppendorf, Germany). The ERIC-PCR reaction was repeated at least twice for each isolate to verify the reproducibility of the assay. The 16S rRNA gene of the recovered isolates was amplified employing primers of a previous study [Reference Milton16] and sequenced by Sanger dideoxy method using commercial sequencing services (Eurofins Ltd., Bangalore, India). The PCR reaction mixture for amplification consisted of 12.5 µl of 2× PCR master mixtures (ThermoFisher Scientific), 1 µl (10 pmol/μl) of each primer (Eurofins, India), 2 µl of DNA template and nuclease-free water to make final volume up to 25 µl. The cycling conditions for PCR consisted of 5 min initial denaturation at 95 °C followed by 35 cycles each of 1 min denaturation at 94 °C, 30 s annealing at 63 °C and 45 s extension at 72 °C and a final extension step of 5 min at 72 °C. The nucleotide sequences were deposited in GenBank using the National Centre for Biotechnology Information (NCBI, Bethesda, MD) Bankit submission tool http://www3.ncbi.nlm.nih.gov.
Results
Salmonella was detected in 10 of 314 (3.1%) wildlife faecal samples by isolation and 18 of 314 (5.7%) by direct PCR assay; one of 26 (3.8%) feed and water samples and five of 30 (16.7%) caretakers stool swabs. Salmonella was more commonly isolated in faecal samples from golden pheasants (25%; 2/8) and leopard (10%; 2/20). The occurrence of Salmonella in different species is presented in Tables 1–3. Out of 10 isolates from wildlife faecal samples 1 isolate (0.8%) was from captive wild ruminants (n = 126), four isolates (4.6%) were from captive wild non-ruminants (n = 86), five isolates (4.9%) were from captive wild birds (n = 102). Prevalence of Salmonella was further analysed as per the sample group and zoo (Tables 4 and 5). Prevalence of Salmonella spp. among different sample group and different zoos were statistically significant (P < 0.05). By serotyping, 6/16 isolates (37.5%) were found to be S. Typhimurium, four isolates (28.5%) were recognised as S. Kentucky, two isolates (14.3%) were identified as S. Enteritidis, two isolates (14.3%) were untypable, and one each isolate (6.2%) were S. Senftenberg and S. Lamberhurst (Table 6).
Table 4. Distribution of Salmonella spp. among the different sample groups
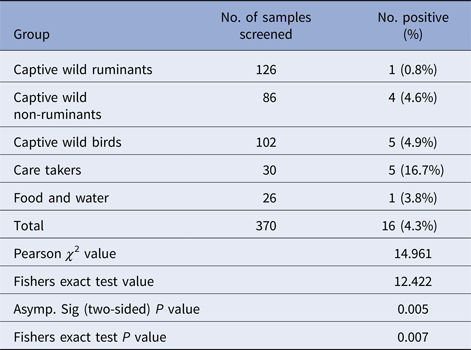
Table 5. Distribution of Salmonella spp. among different zoos/enclosures
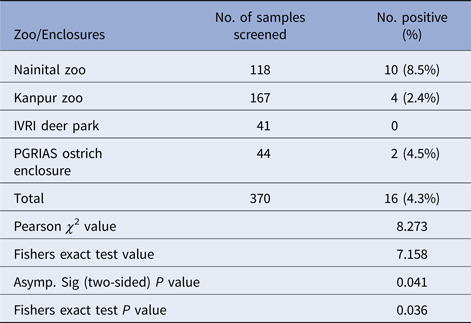
Table 6. Characterisation of Salmonella isolated from this study
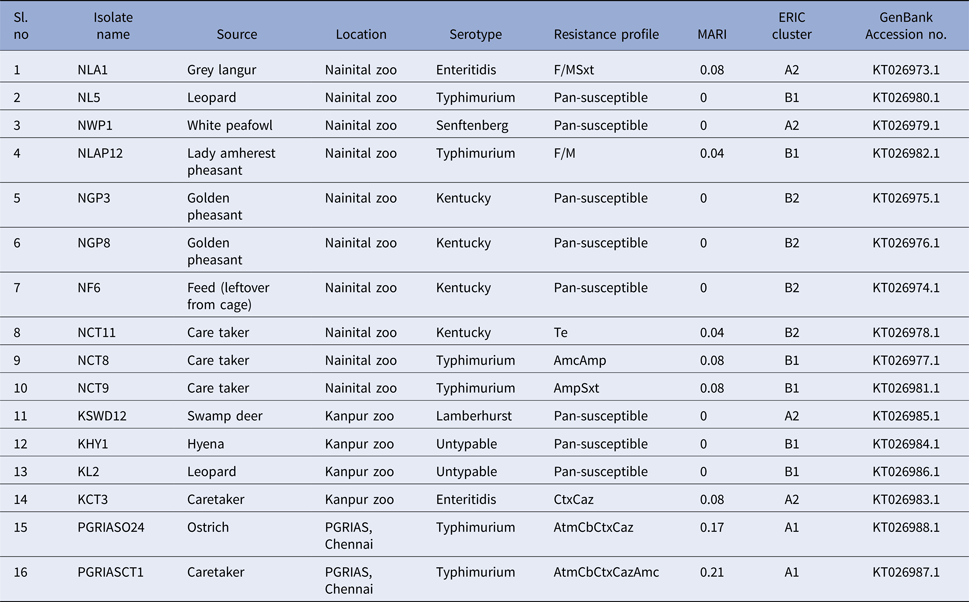
We found antimicrobial resistance among the isolates at varied frequencies. The highest frequency of resistance was found against cefotaxime (3; 18.7%) and ceftazidime (3; 18.7%), followed by carbenicillin (2; 12.5%), aztreonam (2; 12.5%), amoxiclav (2; 12.5%), sulphamethoxazole with trimethoprim (2; 12.5%), nitrofurantoin (2; 12.5%), ampicillin (2; 12.5%) and tetracycline (1; 6.2%). No resistance was found against streptomycin, gentamicin, kanamycin, amikacin, colistin, ceftazidime + clavulanic acid, cefotaxime + clavulanic acid, erythromycin, enrofloxacin, ciprofloxacin, ofloxacin, meropenem, imipenem, ertapenem. The overall multidrug resistance was low (2/16; 12.5%). Seven (7/16; 43.7%) of the isolates were resistant to one antibiotic and one isolate each exhibited penta and tetra resistance MDR with AtmCbCtxCazAmc and AtmCbCtxCaz R-types, respectively. It should be noted that these two isolates belonged to serovar Typhimurium and were recovered from the ostrich and its caretaker. Eight of the isolates (50%) were pan-susceptible to the panel of 23 antimicrobials included in this study. These isolates belonged to Kentucky (n = 3), Typhimurium (n = 1), Senftenberg (n = 1), Lamberhurst (n = 1) and Untypable serovars (n = 2). MARI among the isolates ranged from 0 to 0.21. Resistance patterns and MARI of the Salmonella serovars are shown in Table 6.
The ERIC PCR typing of 16 S. enterica isolates to determine the genetic diversity and phylogenetic relationship among the strains yielded amplified fragments of size ranging from 150 to 2500 bp and were distributed in two clusters (A & B) with two subclusters each with a Simpson's discriminative index of 0.867 (Fig. 1). Six and ten isolates were grouped in clusters A and B, respectively. Four S. Kentucky isolates from two golden pheasants, feed and its caretaker of Nainital zoo grouped into the same subcluster B2 of cluster B. Similarly S. Typhimurium isolates from ostrich and its caretaker of Chennai grouped into same subcluster A1 of cluster A. Two untypable Salmonella isolates from a hyena and leopard of Kanpur zoo grouped with a S. Typhimurium isolate from a leopard of Nainital zoo in a same subcluster B1 of cluster B. Three S. Typhimurium isolates from two caretakers and a lady Amherest pheasant of Nainital zoo grouped in a same subcluster B1 of cluster B. S. Enteritidis from grey langur and S. Senftenberg from white peafowl of Nainital zoo grouped in a subcluster A2 of cluster A. Similarly S. Enteritidis isolated from caretaker of Kanpur zoo grouped with S. Lamberhurst isolated from swamp deer of Kanpur zoo in a subcluster A2 of cluster A. These results coincided with the 16S rRNA gene sequences submitted in GenBank and the accession numbers are indicated in Table 6.
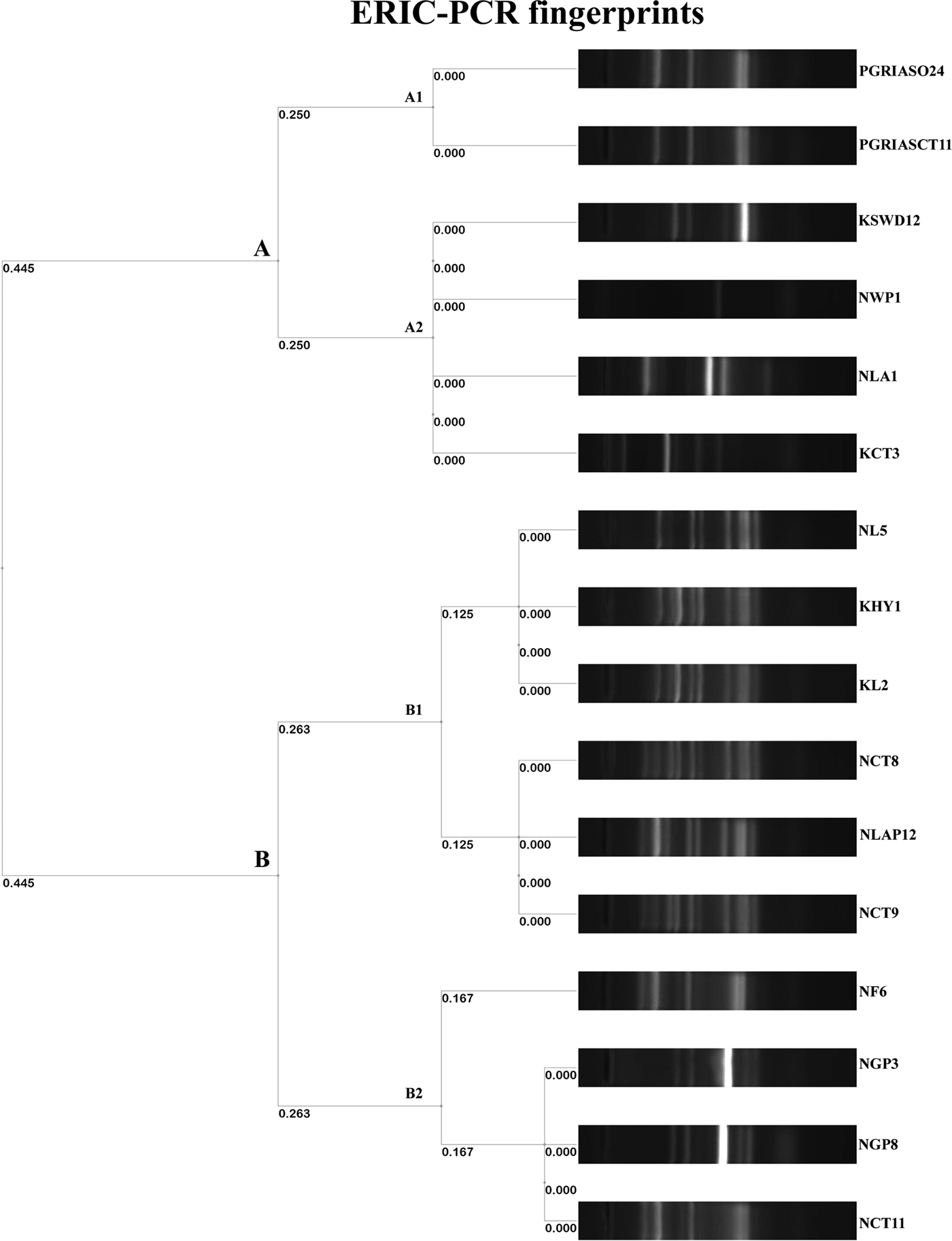
Fig. 1. Dendrogram representing genetic relationships among Salmonella strains based on ERIC-PCR fingerprints.
Discussion
The present study documents the occurrence of Salmonella among captive wildlife in India for the first time and indicates the importance of captive wildlife as potential sources of human infections through occupational or other direct or indirect contact with wild animals. In this study, the prevalence varied among the host species (Tables 1–3). The number of tested animals is low for several species, because of the lesser exhibits kept in a zoo. This is the first report regarding the prevalence of Salmonella in healthy captive wild animals in India except for few case reports [Reference Mahajan17–Reference Shilpa19]. Previous studies from other countries like the USA have shown that free-range wildlife species such as wild pig, deer, opossum, coyote, crow, elk, etc. could be the main source of S. enterica contamination to water, cattle, pre-harvest lettuce and spinach [Reference Gorski3]. To the best of our knowledge and based on available literature, isolation of Salmonella from swamp deer, lady Amherst pheasant and white peafowl appears to be for the first time in the world.
In previous studies Salmonella was isolated from grey langur [Reference Parmar20], hyena [Reference Gitter and Brand21], leopard [Reference Clyde22], golden pheasants [Reference Ahasan and Rahaman23] and ostrich [Reference de Freitas Neto24]. The isolation rate of Salmonella from the feces of all captive wildlife was 3.1% and by direct PCR assay, detection rate was found to be 5.7%. Direct PCR detection of Salmonella from feces has been reported in many previous studies [Reference Cohen25–Reference Soria28]. Our results were in accordance with the previous studies where PCR was found more sensitive than the culture methods. Rychlik et al. [Reference Rychlik29] reported that the sensitivity of the culture method may be lower than that of DNA-based detection assays because of the inability of culture to detect sublethally injured or viable non-culturable cells. The predominant serovars identified in this study were Typhimurium and Kentucky. Both of these serovars were identified earlier from human infections in India [Reference Ballal30]. Two Typhimurium strains isolated in this study were MDR with penta and tetra-type resistant patterns suggesting the significance of this serovar in animal and public health. MDR S. Typhimurium which is associated with invasive infections and mortality in humans has previously been reported from human, domestic and wild animals [Reference Singh31, Reference Molina-Lopez32]. In our study, majority of the isolates were pan-susceptible, which is found to be in accordance with a previous study on wildlife [Reference Farias6] and is probably linked to less usage of antibiotics in zoos compared with animal farms. It is clear from our study, prevalence of animal faecal carriers are low. The route of Salmonella transmission are multifaceted and our findings suggest that environmental contamination through indirect sources such as infected animal or human faeces and feed could play important roles. Nevertheless direct transmission of Salmonella from captive wildlife to visitors or caretakers is rare. Genotyping of isolates in the current study showed that most of the Salmonella isolates within serovar are clonally related (Fig. 1). Interestingly, clonally related isolates detected in both animals and caretakers suggest the role of wildlife in transmission, even if it is difficult to determine its direction. In addition, the presence of clonally related isolates in different species of captive wild animals from the same zoo shows that the spread could be due to fomites including caretakers, visitors, vehicles, other implements and other animal species, such as rodents and wild birds. Further we suggest intensive longitudinal studies which may also shed light on various risk factors involved. In conclusion, our study suggests that captive wildlife is asymptomatic carriers of Salmonella. The point to be noted is prevalence of shedding in animals was low. The occurrence of Salmonella and different serovars in captive wildlife species and resistance of some isolates are of public health concern. Understanding the epidemiology and transmission pattern of Salmonella between captive wildlife and caretakers could help to prevent and control the introduction and spread of infections among people.
Acknowledgements
We would like to express our gratitude to ICAR for funding the project, Outreach Programme on Zoonoses.
Conflict of interest
None.
Ethical standards
Not required.









