INTRODUCTION
Group B Streptococcus (GBS), or Streptococcus agalactiae, frequently colonise the genitourinary tract in pregnant women and can cause invasive disease in neonates, with fatality rates as high as 10% [Reference Edmond1]. Up to 25% of neonates that survive infection may suffer severe neurological impairment [Reference Libster2]. In recent years, GBS has also emerged as an important pathogen in non-pregnant adults, particularly the elderly and those with underlying diseases such as diabetes [Reference Lamagni3].
Ten serotypes of GBS exist but studies have shown that serotypes Ia, Ib, II, III and V occur more frequently and are largely responsible for invasive disease [Reference Edmond1, Reference Lamagni3]. GBS are further classified by sequence types (ST) based on multilocus sequence typing (MLST), with ST-1, ST-10, ST-17, ST-19 and ST-23 being more frequently associated with invasive infection [Reference Edmond1, Reference Jones4, Reference Meehan, Cunney and Cafferkey5]. The ‘hypervirulent’ clone ST-17 is often linked with serotype III strains in invasive neonatal disease [Reference Tazi6] but recent studies have also shown an association between serotype V, ST-1 strains and invasive disease in non-pregnant adults [Reference Flores7].
Current practice in Ireland for prevention and control of neonatal GBS disease involves a risk-based assessment, as per the Royal College of Obstetricians and Gynaecologists guidelines, with intrapartum antibiotic prophylaxis (IAP) administered only to those considered at significant risk [8]. However, the use of IAP has raised concerns regarding the potential for development of antibiotic resistance in GBS isolates [Reference Phares9]. Penicillin remains the first line agent for IAP and treatment of GBS infections [Reference Verani, McGee and Schrag10], however, isolates with reduced susceptibility have been confirmed in some countries [Reference Kimura11]. When a patient presents with beta-lactam allergies, alternative treatment options are required, with lincosamides (clindamycin) and macrolides (erythromycin) considered the next choice of treatment, but resistance levels to these antibiotics continue to rise [Reference Lamagni3, Reference Verani, McGee and Schrag10].
Macrolide resistance is mediated by two mechanisms with ribosomal modification by methylation (encoded by erm genes) being the most common, and efflux pumps. In streptococci, ermB and ermTR genes confer resistance to erythromycin, and constitutive or inducible resistance to clindamycin and streptogramin B antibiotics (c- or i-MLSB phenotypes respectively). Efflux pumps are encoded by mef genes and restrict resistance to 14- and 15-membered macrolides only (e.g. erythromycin), which is associated with the M phenotype [Reference Roberts12, Reference Leclercq13].
Resistance to clindamycin occurs primarily through erm genes, however, isolates displaying resistance to clindamycin but susceptibility to erythromycin (L phenotype) also occur. This unusual mechanism of lincosamide-specific resistance occurs by enzymatic inactivation of the drug by nucleotidyl-transferases and is mediated by lnu genes [Reference Bozdogan14]. Furthermore, this type of lincosamide resistance may also occur with cross resistance to streptogramins A and pleuromutilins, designated LSA and LSAP phenotypes, which are mediated by the ABC transporter, lsaC [Reference Malbruny15]. A retrospective study conducted at the Centers for Disease Control and Prevention in 2016 confirmed that while the incidence of these rare phenotypes is low, their rates are increasing and thus should be actively monitored [Reference Hawkins16].
This study aimed to provide current epidemiological data on serotype distribution and antibiotic resistance rates among colonising and invasive GBS isolates in the South of Ireland, and to investigate the genetic basis for antibiotic resistance and the emergence of new strain phenotypes.
MATERIALS AND METHODS
Bacterial isolates
A total of 235 GBS isolates collected over a 6-year period (2010–2016) were included in this study. Colonising isolates (n = 201) were collected from the Microbiology Department at Cork University Hospital, obtained from high vaginal swabs (HVS) of women between the ages of 16 and 45 years; one colonising GBS isolate was recovered from a placental swab (PS) and was provided by the National Maternity Hospital, Holles Street, Dublin. Invasive GBS isolates (n = 34) were collected from blood culture samples from Cork University Hospital and University Hospital Limerick. Isolates were cultured on Columbia Blood Agar (Fannin Ltd, Dublin, Ireland) and incubated at 37 °C for 24 h. Isolates were identified as GBS by beta-haemolysis on blood agar, a positive Gram stain and a negative catalase test and identified to species level by MALDI-TOF MS (Bruker Daltonics, Leipzig, Germany).
Antimicrobial susceptibility testing
Antimicrobial susceptibility of the isolates was assessed by a disk diffusion method, according to EUCAST guidelines (http://www.eucast.org/ast_of_bacteria/) and all isolates were tested for susceptibility to penicillin, erythromycin, clindamycin, rifampicin and tetracycline. Inducible resistance to clindamycin was determined using the double-disk diffusion method (D test) (EUCAST) and resistance phenotypes were characterised as constitutive (c-MLSB), inducible (i-MLSB), M phenotype (erythromycin resistant, clindamycin sensitive) or L phenotype (erythromycin sensitive, clindamycin resistant). Minimum inhibitory concentration (MIC) testing of erythromycin and clindamycin was performed using MIC Evaluator strips (Oxoid, Basingstoke, UK) and interpreted according to EUCAST breakpoints (http://www.eucast.org/clinical_breakpoints/).
DNA extraction from bacterial cultures
DNA was extracted using the PureLink Genomic DNA Kit (Invitrogen, ThermoFisher Scientific, California, USA) with minor modifications, which included a mechanical lysis of GBS by bead-beating using the MagNA DNA lyser (Roche, Indiana, USA). Briefly, an overnight culture of GBS in Brain Heat Infusion broth (BHI, Cruinn Diagnostics, Dublin, Ireland) was centrifuged at 6500 rpm for 10 min. The supernatant was removed and the cell pellet was re-suspended in 1 ml TE buffer. To the washed cells, 470 mg of 150–212 µm glass beads (Sigma-Aldrich, Missouri, USA) were added and the samples were bead-beated at 6500 rpm for 60 s. Samples were then centrifuged at 13 000 rpm for 10 min. From each lysed sample, 240 µl of supernatant was removed and processed using the extraction and purification protocol described by the manufacturer, beginning with the proteinase K digestion step.
Molecular characterisation of antimicrobial resistance genes
A multiplex PCR assay was used to detect the presence of the erythromycin resistance genes ermB and mefA/E, with inclusion of an S. agalactiae-specific internal control targeting the cfb gene [Reference Sutcliffe17, Reference Ke18]. Other genes involved in promoting erythromycin resistance (ermTR and ermT) were detected by PCR as previously described [Reference De Azavedo19, Reference Compain20]. A multiplex assay was performed to detect the clindamycin resistance genes lsaC and lnuB [Reference Bozdogan14, Reference Malbruny15]. Additional clindamycin resistance genetic determinants (lnuA, lsaB, lsaE and vgaC) were investigated by PCR according to previous studies [Reference Arana21, Reference Douarre22].
Capsular typing and ST-17 detection
All isolates were assigned serotypes according to the recommended polymerase chain reaction (PCR)-based capsular genotype algorithm [Reference Yao23]. Briefly, isolates were initially serotyped using two multiplex PCR reactions [Reference Poyart24] and any serotype VII or non-typeable isolates were further identified by PCR [Reference Imperi25]. Detection of the hypervirulent ST-17 clone was performed using a modified version of a multiplex PCR assay targeting a region of the hvgA gene [Reference Gosiewski, Brzychczy-Włoch and Heczko26].
Statistical analysis
The χ 2 test and Fisher's exact test were used to analyse associations using SPSS v. 24·0 (IBM, UK). A P-value ⩽0·05 was considered statistically significant.
RESULTS
Antimicrobial susceptibility
All isolates (n = 235) were susceptible to penicillin and rifampicin. Resistance to tetracycline, erythromycin and clindamycin (both constitutive and inducible) was observed in 87·2% (n = 205), 21·3% (n = 50) and 20·4% (n = 48) of isolates, respectively. Differences in overall resistance rates were evident between invasive (n = 34) and colonising (n = 201) isolates, with all of the former group being resistant to tetracycline compared with 85·1% (n = 171) of colonisers. Similarly, erythromycin resistance was found in 35·3% (n = 12) of invasive and 18·9% (n = 38) of colonising isolates (P = 0·041), and clindamycin resistance in 32·4% (n = 11) and 18·4% (n = 37) of invasive and colonising isolates, respectively (P = 0·069).
Macrolide resistance phenotypes and genotypes
In total, 50 isolates were resistant to erythromycin (n = 12 invasive; n = 38 colonising) and of these, 62% (n = 31) exhibited the c-MLSB phenotype (n = 9 invasive; n = 22 colonising), while 20% (n = 10) were of the i-MLSB phenotype (n = 2 invasive; n = 8 colonising) and 18% (n = 9) belonged to the M phenotype (n = 1 invasive, n = 8 colonising) (Table 1). The ermB gene was the most frequent (84%, n = 42) among macrolide resistant isolates while mefA/E and ermTR genes were each detected in 18% (n = 9) of isolates; ermT was restricted to a single isolate, which was also positive for the ermB gene. One isolate harboured a combination of ermB and mefA/E genes, and another was positive for both ermTR and mefA/E. All 31 isolates exhibiting the c-MLSB phenotype were ermB positive and three of these (9·7%) also harboured the ermTR gene. The ermB gene was detected in all 10 isolates displaying the i-MLSB phenotype and five of these isolates were positive for the ermTR gene; another single isolate harboured the ermT gene. All M phenotype (n = 9) representatives contained the mefA/E resistance determinant, with only one isolate being additionally positive for ermB (Table 1).
Table 1. Genotypic and phenotypic characterisation of group B streptococcal isolates resistant to erythromycin
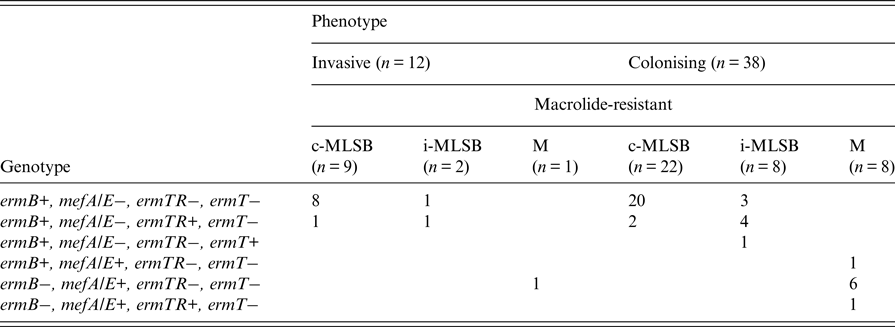
c-MLSB, constitutive macrolide and lincosamide resistance; i-MLSB, inducible clindamycin resistance in the presence of erythromycin; M, resistance to erythromycin only.
Characterisaton of the L phenotype
The L phenotype of erythromycin sensitivity and clindamycin resistance was identified in 2·9% (n = 7) of all GBS isolates and confirmed by MIC susceptibility testing (Table 2). Four isolates were positive for the the lsaC gene and the remaining three were negative for all genetic deteminants investigated. The L phenotype was not detected among invasive isolates.
Table 2. Characterisation of L phenotype isolates of group B streptococci
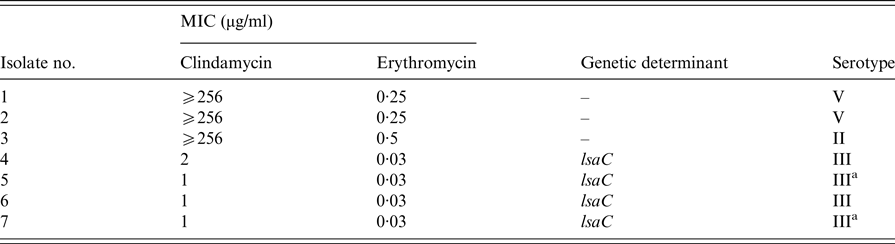
All isolates were screened for the presence of lnuA, lnuB, lsaB, lsaC, lsaE and vgaC determinants. Isolates 1–6 were of high vaginal swab origin, isolate 7 was from a placental swab.
a Isolates of the ST-17 clone.
Serotype distribution amongst GBS populations
Nine serotypes were identified among the total study collection; serotype IX was not found and one isolate was non-typeable by both PCR methods. The most common serotypes were Ia (28·1%), III (24·7%), II (14%), V (12·8%) and Ib (11·9%), and serotypes IV, VI, VII and VIII accounted for 4·3%, 1·3%, 1·7%, and 0·85% of isolates, respectively. Among colonising isolates the predominant serotypes were serotypes Ia (29·4%), III (25·9%), II (15·4%), V (11·4%) and Ib (9·95%). However, the serotype distribution for invasive isolates differed, with Ib (23·5%), Ia (20·6%), V (20·6%) and III (17·6%) being the most prevalent, but the difference between the two isolate groups did not reach statistical significance (P = 0·081).
The hyper-virulent clone, ST-17, was identified in 12·3% (n = 29) of all isolates, with 20·6% (n = 7) and 10·9% (n = 22) accounting for invasive and colonising populations. All, but one, of the ST-17 isolates were serotype III (P < 0·0005), the other isolate being confirmed by both PCR methods as serotype IV.
Table 3 shows that there was no significant difference in serotype distribution between neonatal and adult invasive isolates (P = 0·378). Two invasive adult isolates were serotype III and ST-17, and another strain of serotype IV/ST-17 was also confirmed from an adult sample. Of the 52 serotype III colonising isolates, 42·3% (n = 22) were ST-17.
Table 3. Correlation of sample origin and serotype distribution amongst invasive and colonising isolates of group B streptococci
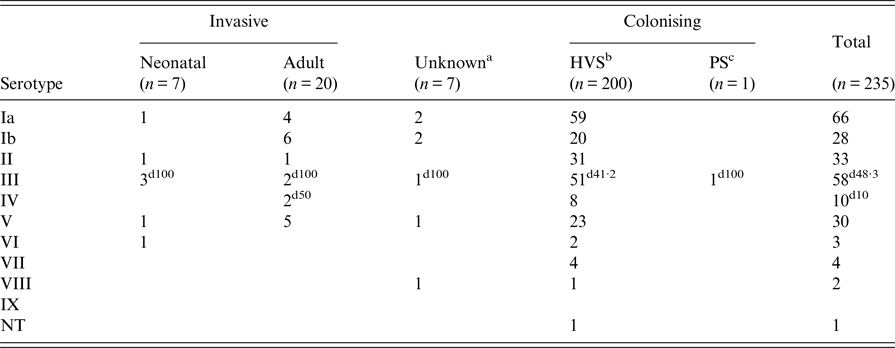
a Lack of clinical information.
b High vaginal swab.
c Placental swab.
d Percentage of ST-17 isolates within group.
Within the 50 isolates resistant to erythromycin, serotype III predominated (n = 15) followed by serotype V (n = 12), Ia (n = 8), II (n = 6) and Ib (n = 4). Both serotypes IV and VII were found in two isolates each and serotype VI was identified in a single isolate only. The majority of the 15 serotype III erythromycin resistant isolates expressed the c-MLSB phenotype (86·7%) and all harboured the ermB gene; the c-MLSB phenotype was also found in 9 of 12 serotype V isolates but these harboured varied erythromycin resistance determinants (Table 4). Significant associations were found between serotype Ia and the M phenotype (P = 0·0001) and serotype Ia and the mefA/E gene (P = 0·00046). Serotype V was also frequent among the invasive erythromycin-resistant population (41·7%), while serotype III predominated among colonising isolates resistant to erythromycin (31·6%) but there was no significant difference in serotype distribution within the two populations (P = 0·317). Three of the 12 erythromycin resistant invasive isolates were ST-17 and were also resistant to clindamycin; correspondingly, three colonising isolates of the resistance phenotype were members of the same ST-17 clone.
Table 4. Correlation between serotype, phenotype and genotype of group B streptococcal isolates
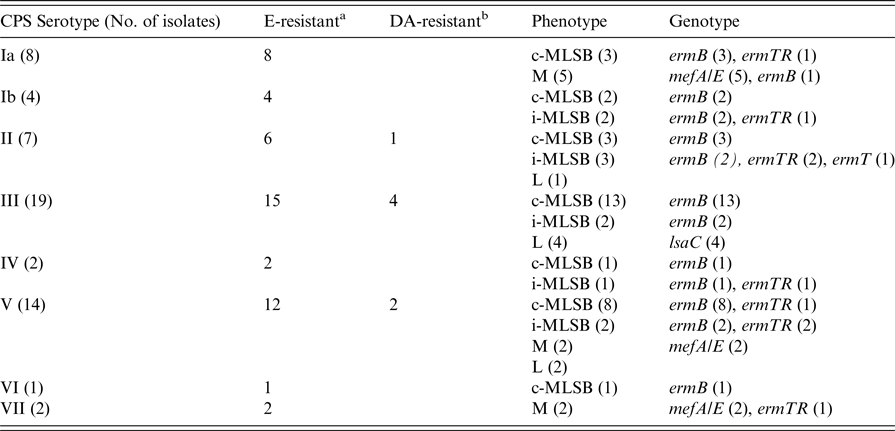
a E-resistant = all erythromycin-resistant isolates resistant or sensitive to clindamycin.
b DA-resistant = isolates resistant to clindamycin but susceptible to erythromycin.
Four of the seven isolates exhibiting the L phenotype were serotype III, two were of serotype V and one was serotype II (Table 2). The four L phenotype serotype III isolates harboured the lsaC gene and two of these were ST-17.
DISCUSSION
Despite recent reports of reduced beta-lactam susceptibility among GBS isolates [Reference Kimura11], data from this study indicate that susceptibility to penicillin is universal amongst GBS strain populations in Ireland. This reaffirms the appropriateness of penicillin as the antibiotic of choice for first line IAP and for the treatment of GBS infections. The incidence of erythromycin resistance found (21·3%) is similar to previous studies conducted in Ireland (18·6%), Poland (18·4%) and Italy (20%) [Reference Meehan, Cunney and Cafferkey5, Reference Sadowy, Matynia and Hryniewicz27, Reference Bezares28], but lower than rates in France (34·7%) and Taiwan (46%) [Reference Bergal29, Reference Hsueh30]. Interestingly the rate of clindamycin resistance in the current study indicates that resistance has increased in the Irish population from 15·25% [Reference Meehan, Cunney and Cafferkey5] to 20·4% and is higher than reports from other countries, including Poland and Africa [Reference Sadowy, Matynia and Hryniewicz27, Reference Belard31]. As with other countries, a high rate of tetracycline resistance (87·2%) was evident for the isolates reported here [Reference Bezares28, Reference Bergal29].
Colonising GBS isolates can be reservoirs of virulence and antibiotic resistance markers and vaginal colonisation may pose a risk for subsequent invasive disease in neonates. Both erythromycin and clindamycin resistance are higher among invasive (35·3% and 32·4% respectively) than colonising isolates (18·9% and 18·4% respectively), although this only reached statistical significance for erythromycin resistance (P = 0·041).
Regarding antibiotic resistance mechanisms, the c-MLSB phenotype predominated overall (62%) and was more frequent among invasive (75%) than colonising isolates (57·9%). The ermB gene was the most common resistance determinant identified (84%) and this finding corroborates the results of the most recent Irish study [Reference Meehan, Cunney and Cafferkey5]. The presence of the ermT gene, although only in a single isolate, is notable as it was associated with the i-MLSB phenotype. This determinant may be of importance as it is plasmid-borne and has been shown to be readily transferable between GBS and Enterococcus faecalis, which could promote its wider dissemination [Reference Compain20]. It is interesting to note the co-occurrence of ermB and ermTR genes in eight isolates and mefA/E and ermTR genes in another isolate. Co-occurrence of ermB and ermTR has also been noted in other studies, although in a lower number of isolates [Reference Bezares28, Reference Bergal29, Reference Seo32, Reference Metcalf33].
The L phenotype was first documented in E. faecium in 1999 [Reference Bozdogan14] and followed shortly after by the first report in a GBS strain in Canada in 2001 [Reference De Azavedo19]. Since then there have been reports of the L phenotype in Spain [Reference Arana21], Korea [Reference Seo32], the USA [Reference Gygax34], Argentina [Reference Faccone35] and South Africa [Reference Bolukaoto36] with incidence rates varying from 0·26% in Italy [Reference Bezares28] to 15·9% in Korea [Reference Seo32]. To date, the phenotype had not been confirmed in GBS isolates from Ireland but was found here to represent almost 3% of isolates which, interestingly, were collected in the latter part of the study period (2015–2016).
Four of the seven L phenotype isolates harboured the lsaC gene and were serotype III. This observation concurs with reports from previous studies [Reference Malbruny15, Reference Hawkins16, Reference Arana21, Reference Seo32, Reference Björnsdóttir37]. Moreover, the three other isolates of this phenotype with an MIC for clindamycin >256 µg/ml were negative for all other determinants tested including the lnu, lsaB, lsaE and vgaC genes. Two of these isolates were confirmed as serotype V and the other was serotype II. To the best of our knowledge, this is the first documented report of GBS isolates with the L phenotype and characterised as containing the lsaC gene in Ireland. Although the L phenotype was exclusively identified in colonising isolates in this study, it has been found by others in patients with invasive GBS disease [Reference Hawkins16, Reference Björnsdóttir37]. Also of interest, two of the seven isolates were confirmed as belonging to the hypervirulent ST-17 clone. The emergence of this uncommon phenotype in Irish GBS strains corroborates the need for continued surveillance of emerging antibiotic resistance, as recently reported in the USA [Reference Hawkins16]. This USA study also revealed the presence of L phenotypes with cross-resistance to streptogramins A and pleuromutilins, the LSA/LSAP phenotypes, which have been reported elsewhere [Reference Malbruny15, Reference Björnsdóttir37]. Another recent study using whole genome sequencing to identify resistance mechanisms in GBS, identified the LSA/LSAP phenotypes using this approach and highlighted the co-occurrence of certain genes, particularly lsaC and ermB, amongst isolates displaying this phenotype [Reference Metcalf33]. While we did not specifically investigate the LSA/LSAP phenotypes, they may be clinically significant going forward and support the need for further study into the development of alternative antimicrobials.
Here, nine serotypes were confirmed among the isolate collection and only one isolate proved non-typeable by both PCR methods used. Overall, serotype Ia predominated (28·1%), followed by types III, II and V. This distribution is comparable with a previous Irish study [Reference Meehan, Cunney and Cafferkey5] and indicates a level of stability within the GBS population in this country. Serotype III was also predominant among erythromycin-resistant isolates in contrast to a previous study in the South of Ireland where serotype V was more frequently associated with erythromycin-resistant strains [Reference Kiely, Lucey and Cotter38].
The ST-17 lineage accounted for 12·3% of all isolates studied with rates of 20·6% and 10·9% in the invasive and colonising populations respectively. The high prevalence of this clone in the latter group is of concern because of its potential to progress to invasive disease, thus highlighting the importance of monitoring reservoirs of colonising strains. Three of the ST-17 invasive isolates were of neonatal origin and of serotype III, and three were from invasive adult infections, but of different serotypes (III and IV) which may suggest the occurrence of capsular antigen switching as reported in other studies [Reference Meehan, Cunney and Cafferkey5, Reference Bellais39]. Together, these findings serve to emphasise the need for continuous surveillance of serotypes which is highly relevant to the development of vaccines [Reference Leroux-Roels40].
In conclusion, the increase in resistance to erythromycin and clindamycin in GBS strains is of significance particularly for individuals with penicillin allergies. Emergence of the L phenotype in the Irish GBS population is a concern and warrants continued surveillance of resistance rates and further elucidation of the underlying genetic mechanisms.
ACKNOWLEDGEMENTS
The authors are extremely grateful to staff at the Microbiology Department of University Hospital Limerick and Cork University Hospital for the provision of GBS isolates. The authors also thank the Microbiology Department of The National Maternity Hospital, Holles Street. This work was supported by the RÍSAM Scholarship Programme of Cork Institute of Technology.
DECLARATION OF INTEREST
The authors declare no conflict of interest.







