The gut microbiome is a rapidly expanding area of human research(Reference Cani1). While often used interchangeably(Reference Knight, Callewaert and Marotz2), the term gut ‘microbiome’ refers to all of the microbial genes that reside inside the gastrointestinal tract, while the microbes themselves are collectively known as the ‘microbiota’. Being a complex and metabolically active ecosystem, these microbes generally maintain a symbiotic relationship with the host and participate in a number of beneficial functions within the body. These include the synthesis of vitamins(Reference LeBlanc, Milani and de Giori3), regulating the immune system(Reference Chu and Mazmanian4) and production of SCFA, which are a key energy source for colonic epithelial cells(Reference Ramakrishna5). Interestingly, several health conditions have been linked with alterations in the gut microbiota populations, including type 2 diabetes(Reference Larsen, Vogensen and van den Berg6,Reference Qin, Li and Cai7) , obesity(Reference Ley, Turnbaugh and Klein8,Reference Turnbaugh, Hamady and Yatsunenko9) and inflammatory bowel disease(Reference Morgan, Tickle and Sokol10); however, it is unclear if this dysbiosis is a cause or consequence of the disease(Reference Fischbach11).
Relatively recent advancements in techniques used to characterise the composition of the microbiota have provided a greater understanding of host–gut microbiota metabolic interactions and its subsequent effect on human physiology. Since 2005, advancements in microbial determination methods, in particular high-throughput sequencing techniques, have vastly improved the understanding of the gut microbiota, by allowing a more comprehensive investigation of phylogenetic composition and quantification(Reference Fraher, O’Toole and Quigley12). Examples of this technology include the next-generation sequencing of the 16s rRNA gene or its amplicons, which are based on sequence divergences of the small subunit rRNA(Reference Allaband, McDonald and Vazquez-Baeza13).
Given the potential impact of the gut microbiome on human health and disease, there is a need to identify foods which may support a healthy microbiome. Previous research has shown that prebiotic fibres are a key substrate facilitating change in the microbiome(Reference So, Whelan and Rossi14). In humans, prebiotic fibre escapes digestion in the small intestine and instead passes into the colon where it is used as a substrate by the microbes. Here, it stimulates the growth of specific organisms leading to the production of different metabolites, including the SCFA butyrate, which confers health benefits to the host(Reference Holscher15).
Nut consumption is associated with many positive health benefits(Reference Ros16). Both epidemiological studies and clinical trials have linked frequent nut consumption to a reduced risk of developing type 2 diabetes(Reference Viguiliouk, Kendall and Blanco Mejia17) and CHD(Reference Kris-Etherton, Hu and Ros18), as well as a reduction in cardiovascular risk factors such as hypertension(Reference Mohammadifard, Salehi-Abargouei and Salas-Salvado19) and the promotion of a more beneficial lipid profile(Reference Griel and Kris-Etherton20). Nuts are nutrient dense and contain high amounts of plant-based protein, unsaturated fatty acids, vitamins, minerals and other phytochemicals(Reference Brufau, Boatella and Rafecas21). Nuts have the second highest fibre content of all foods per 100 g, behind cereals(Reference Marlett22). Nuts are also a food source rich in polymerised polyphenols(Reference Lamuel-Raventos and Onge23), resistant starch(Reference Fuentes-Zaragoza, Sánchez-Zapata and Sendra24) and NSP(Reference Sargautiene, Nakurte and Nikolajeva25), which appear to have a prebiotic effect. Additionally, walnuts are particularly high in n-3 fatty acids, with preliminary research classifying these fatty acids as prebiotics(Reference Costantini, Molinari and Farinon26). While the health benefits of nuts may in part be due to their unique nutritional composition, the exact mechanism by which nuts exert this range of beneficial health effects remains unclear. Previous trials have examined the specific microbial shifts that occur with nut consumption(Reference Holscher, Guetterman and Swanson27–Reference Ukhanova, Wang and Baer34); however, due to inconsistency of study findings, there is a need to evaluate the evidence base. The present systematic review aimed to synthesise the existing evidence regarding the effect of nut consumption (tree nuts and peanuts) on the gut microbiota of humans. We hypothesise that due to the array of prebiotic compounds, supplementation of the diet with nuts promotes the proliferation of beneficial microbial species, leading to favourable changes in the gut microbiota.
Methods
This systematic review was reported according to the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analysis statement(Reference Moher, Liberati and Tetzlaff35). The review protocol was prospectively registered in PROSPERO (International Prospective Register of Systematic Reviews) protocol no. CRD42019127318.
Literature search
A systematic search of the electronic databases MEDLINE (EBSCO), PubMed, Cochrane CENTRAL and CINAHL (EBSCO) was performed until 28 November 2019, using a combination of MeSH and individual search terms. No date or language restriction was applied in the search strategy. In conjunction with searching the electronic databases, additional hand searching of the reference lists of relevant articles was also performed. An example search strategy is presented in online Supplementary Data 1.
Study selection
All search results were exported from the electronic databases into the reference management software ENDNOTE (X9; Thomson Reuters), and duplicates were then removed. Eligible studies were identified by two review authors (EF and EN) who screened articles independently based on title and abstract. Full texts were then retrieved, and the eligibility criteria were applied to the full-text articles. Discrepancies were resolved by consensus or consultation with a third party (KL).
Studies were included if they met all of the following criteria: (1) human studies involving participants aged more than 3 years old. This broad age range was used due to findings showing that the gut microbiota of children resembles that of an adult after 3 years of age(Reference Yatsunenko, Rey and Manary36); (2) nuts were administered in their whole form or minimally processed to still contain all components of the whole nut (e.g. nut butters, roasted, ground or chopped nuts); (3) consumption of nuts was compared with no consumption of nuts or a lower quantity of nuts; (4) study design was in the form of randomised or non-randomised experiments, cross-sectional or cohort studies and (5) outcomes were measured using next-generation DNA sequencing technologies, including both targeted amplicon sequencing and shotgun metagenomics. Studies were restricted to those that used these methods as they provide the most comprehensive analysis of the microbiota, enabling taxonomic quantification and identification. Additionally, limiting to these methods provided a means of promoting homogeneity between study results, considering the range of microbiota characterisation techniques available and the subsequent differences in reporting of outcome measurements(Reference Fraher, O’Toole and Quigley12).
In addition to the inclusion criteria outlined above, the following exclusion criteria were applied: (1) in vitro experiments and animal studies due to the limited applicability of these results to the human microbiota, (2) experiments where nut intake could not be isolated from other interventions (e.g. other dietary interventions) and (3) studies which only measured changes in the microbiota-related metabolites. In addition, studies which included both nuts in their whole form and isolated nut components (such as oils or extracts) were only included if data from nuts in their whole form could be differentiated.
Outcome measures
Primary outcome measures were between-group differences in α- and β-diversities, as well as statistically significant differences in the taxonomic composition and microbial abundances at specific taxonomic levels (phylum, class, order, family, genus or ‘operational taxonomic units’ (OTU). α-Diversity is a measure of within-sample (or community) diversity, taking into account the richness and/or evenness of the microbes present(Reference Lozupone and Knight37). β-Diversity is a measure of microbial dissimilarity between samples or sites, describing how many taxa are shared between samples(Reference Morgan and Huttenhower38).
Data extraction and quality assessment
A tabular summary was developed for the data extraction process, which included the population, study design and duration, nut type format and dosage, method of microbiota determination and results. Only significant findings (P < 0·05) from each study reporting changes in microbial composition following nut intake were recorded. The quality of included studies was assessed using the Quality Criteria Checklist and Risk of Bias Assessment Tool by the Academy of Nutrition and Dietetics(39).
Results
Summary of included studies
Searches of the electronic databases returned 894 articles, of which fifty-six were evaluated after removal of duplicates and screening of records. A final eight studies met the eligibility criteria and were included in the present review. The above study selection process is summarised in the Preferred Reporting Items for Systematic Reviews and Meta-Analysis 2009 Flow Diagram (Fig. 1) with the included studies summarised in Table 1 and described below. For all studies, significant shifts (P < 0·05) in microbial composition following nut intake are summarised in Table 2.
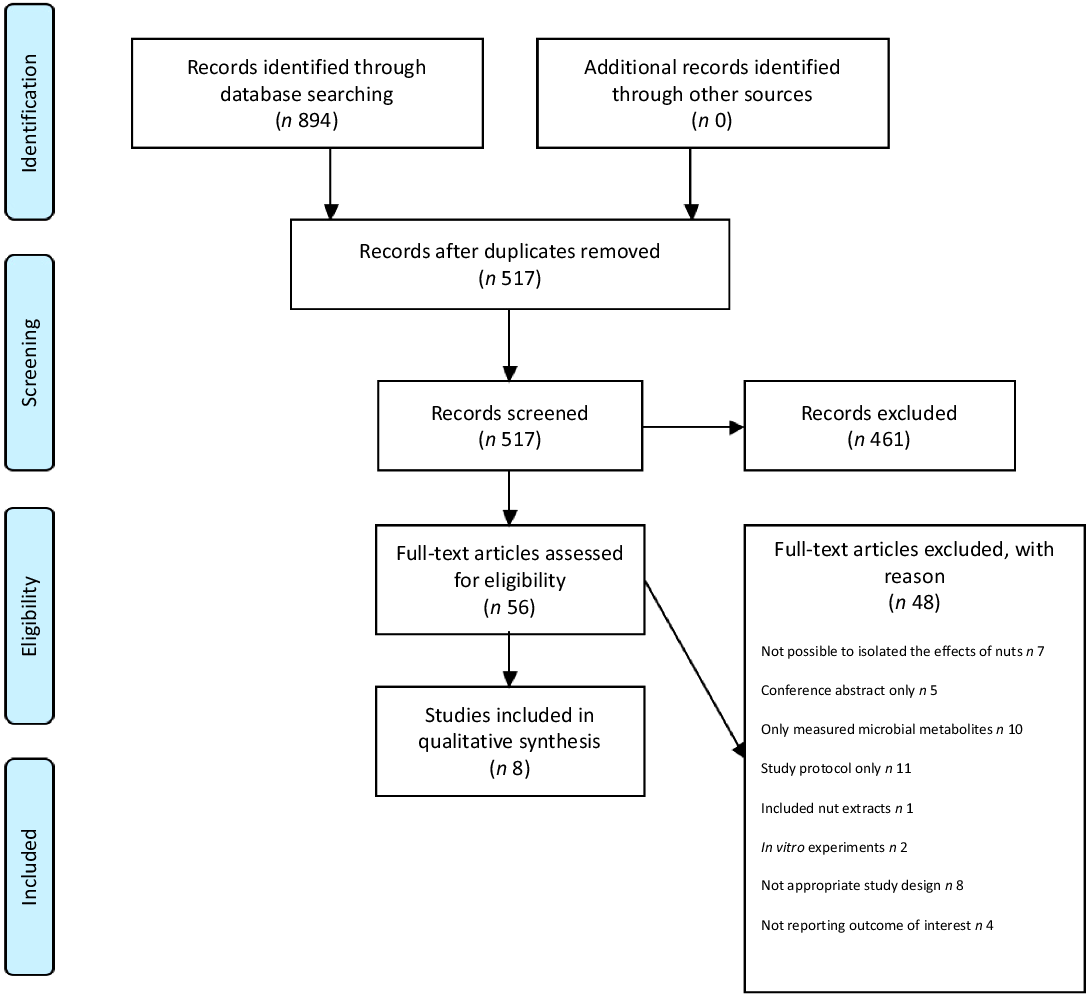
Fig. 1. Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow diagram of study selection.
Table 1. Characteristics of included trials examining the effect of nut consumption on the gut microbiota

RCT-C, randomised controlled trial (cross-over); NHMRC, National Health and Medical Research Council; NR, not reported; PRO, protein; CHO, carbohydrate; OTU, operational taxonomic unit; PCoA, principal coordinate analysis; RCT, randomised controlled trial; UM, urolithin metabotype.
* ‘Feeding trial’ as described above relates to trials in which 100 % of all food consumed during the study duration is provided to participants.
† This study also used fifteen age-matched normolipidaemic controls which were not included in the analysis.
Table 2. Significant shifts in the gut microbial composition following nut intake

↓, Reported decrease in abundance/relative abundance; ↑, reported increase in abundance/relative abundance; OTU, operational taxonomic unit.
† Results for UM-A only.
‡ Results for UM-B only.
Five of the eight studies were randomised controlled cross-over trials(Reference Holscher, Guetterman and Swanson27,Reference Bamberger, Rossmeier and Lechner28,Reference Holscher, Taylor and Swanson31,Reference Burns, Zitt and Rowe32,Reference Ukhanova, Wang and Baer34) with the population group being healthy adults, one of which also included healthy children(Reference Burns, Zitt and Rowe32). All of these studies utilised a washout period, ranging from 1 to 6 weeks, in between treatment periods. The remaining three studies consisted of a parallel design randomised controlled trial(Reference Dhillon, Li and Ortiz33) and two pre-test/post-test studies(Reference Garcia-Mantrana, Calatayud and Romo-Vaquero29,Reference Gargari, Deon and Taverniti30) . Studies were predominantly published in the USA(Reference Holscher, Guetterman and Swanson27,Reference Holscher, Taylor and Swanson31–Reference Ukhanova, Wang and Baer34) (n 5/8), with one being published in Germany(Reference Bamberger, Rossmeier and Lechner28), one in Italy(Reference Gargari, Deon and Taverniti30) and one in Spain(Reference Garcia-Mantrana, Calatayud and Romo-Vaquero29). Sample sizes ranged from n 15 to 194, with the proportion of female participants in each study ranging from 28 to 83 %. Feeding periods within studies ranged from 3 d to 8 weeks, with overall study length ranging from 3 d up to 24 weeks. Three studies examined the effect of whole walnut consumption on the gut microbiota(Reference Holscher, Guetterman and Swanson27–Reference Garcia-Mantrana, Calatayud and Romo-Vaquero29), one study utilised whole roasted hazelnuts(Reference Gargari, Deon and Taverniti30), three studies(Reference Holscher, Taylor and Swanson31–Reference Dhillon, Li and Ortiz33) investigated almonds in varying forms of the whole nut and one study utilised both whole almonds or pistachios(Reference Ukhanova, Wang and Baer34). Nut dose in adult participants varied from approximately one serving (33 g/d) up to two servings daily (84 g), with the dosage being adapted for children. In most studies, the rationale for nut dosage was due to this serving size (42 or 43 g, or 1·5 oz) being consistent with the US Food and Drug Administration qualified health claim for nuts and CVD(Reference Taylor40).
All eligible studies assessed the gut microbiota using high-throughput sequencing technology, with all studies sequencing the 16s rRNA gene to determine bacterial composition. Seven studies used the Illumina Miseq(Reference Holscher, Guetterman and Swanson27–Reference Dhillon, Li and Ortiz33), whereas one used 454-based pyrosequencing platform(Reference Ukhanova, Wang and Baer34). In addition, three studies reported on microbial communities other than bacteria with one study also sequencing fungal DNA(Reference Ukhanova, Wang and Baer34) (using primers directed against the IS4 and IS5 inter-spacer regions), and two studies sequencing both fungal (ITS1–ITS4) and archaeal (16s rRNA gene using 349f/806r primers) DNA(Reference Holscher, Guetterman and Swanson27,Reference Holscher, Taylor and Swanson31) . All studies measured α-diversity using one or a combination of indices (Simpson’s index, Shannon index, Chao-1 index, Faith’s phylogenic diversity or observed OTU). Reported measures of β-diversity used across included studies were the UniFrac and Bray-Curtis distance metrics.
Using the Academy of Nutrition and Dietetics Evidence Analysis Manual Quality Criteria Checklist, six of the eight studies were deemed to be of positive quality(Reference Holscher, Guetterman and Swanson27,Reference Bamberger, Rossmeier and Lechner28,Reference Gargari, Deon and Taverniti30,Reference Holscher, Taylor and Swanson31,Reference Dhillon, Li and Ortiz33,Reference Ukhanova, Wang and Baer34) with the remaining two studies(Reference Garcia-Mantrana, Calatayud and Romo-Vaquero29,Reference Burns, Zitt and Rowe32) found to be of neutral quality (online Supplementary Data 2).
Microbiota diversity metrics and bacterial composition, by nut type
Walnuts
Following consumption of 42 g of walnuts per d for 3 weeks, Holscher et al.(Reference Holscher, Guetterman and Swanson27) found no significant shifts in α-diversity, a measure of within-sample diversity; however, bacterial communities were significantly affected by walnut consumption when β-diversity, a measure of microbial dissimilarity between samples was analysed, as evidenced by a weighted principal coordinate analysis of UniFrac distances (P = 0·03). No significant differences in fungal or archaeal abundances were found following walnut intake. A significant decrease in the relative abundance of Actinobacteria was observed, with the relative abundance of Firmicutes significantly increasing. At the genus level, an increased relative abundance of the genera Faecalibacterium, Clostridium, Roseburia (Clostridium clusters XIVa and IV) and Dialister (49–160 % higher relative abundance) was found, whereas the relative abundances of the genera Ruminococcus, Dorea, Oscillospira and Bifidobacterium were significantly decreased (16–38 % lower relative abundance).
Utilising 43 g of walnuts per d over an 8-week period, Bamberger et al. (Reference Bamberger, Rossmeier and Lechner28) reported a significant shift in β-diversity only. Using the generalised UniFrac distance metric and the PERMANOVA statistical test, significant dissimilarity of approximately 5 % between the walnut and control groups (P = 0·02) was found. No significant differences in predominant phyla or the Firmicutes:Bacteroidetes ratio were observed. Significant shifts in the relative abundance could be seen in four OTUs, which are clusters of similar sequences used to categorise bacteria, of the phyla Firmicutes and one OTU of the phyla Actinobacteria, which decreased. A significant increase was found in two unclassified OTUs from the Ruminococcaceae family (P < 0·02), as well as an increase in an OTU from the Bifidobacterium genus (P < 0·02). A significant decrease was found in the relative abundance of two OTUs, Anaerostipes (P < 0·01) and Blautia (P = 0·04) from the Lachnospiraceae family.
Garcia-Mantrana et al.(Reference Garcia-Mantrana, Calatayud and Romo-Vaquero29) examined the effects of 33 g walnuts per d for 3 d on the gut microbiota, reporting results considering all participants, as well as according to the urolithin metabotype (UM) of participants, a measure of how ellagitannins metabolise to urolithins via the gut microbiota. According to their urinary urolithin concentrations, participants were classified as either UM-A (characterised by the production of urolithin-A), UM-B (characterised by the production of urolithin-B) or UM-0, whereby no urolithins were produced. Local diversity (Chao-1 index) significantly decreased for both UMs after walnut consumption and a redundancy analysis of all individuals revealed that bacterial communities were significantly affected by walnut intake (P = 0·001). However, when results were reported according to UM, these shifts were only significant for UM-B (P = 0·017), suggesting that UM-B was likely a driver of overall significance. Considering the microbial composition, walnut intake significantly decreased the relative abundance of Bacteroidetes, while increasing the abundance of Actinobacteria. Participants classed as UM-A exhibited a significant decrease in the Lachnospiraceae family, as well as a significant increase in the genera Coprococcus and Collinsela. Participants classed as UM-B exhibited a significant increase in the Coriobacteriaceae family, as well as significant increases in the genera Coprococcus, Collinsella, Bifidobacterium and Blautia.
Almonds
Dhillon et al.(Reference Dhillon, Li and Ortiz33) examined the effects of 57 g roasted almonds intake on the gut microbiota over 6 weeks, with graham crackers being given to the control group. When considering α-diversity, almond feeding resulted in significant increases in the Chao-1 index, observed OTUs and Shannon index, while the Simpson’s index was significantly decreased. Similarly, significant differences in β-diversity between treatment groups were identified according to unweighted UniFrac distance and Bray-Curtis dissimilarity metrics. A significant increase was found in bacteria belonging to the RF39 order, as well as a significant decrease in bacteria belonging to the S24.7 family. At the genus level, a significant decrease in Alistipes, Butyricimonas and Odoribacter was observed, with a significant increase in the genera Lachnospira. At the species level, a 48 % decreased relative abundance of Bacteroides fragilis was reported.
Holscher et al.(Reference Holscher, Taylor and Swanson31) utilised 42 g of almonds per d in varying forms over five 3-week diet periods (whole, raw, whole dry roasted, chopped, dry roasted and dry roasted almond butter), finding no significant shifts in microbial diversity measures. Further, no significant differences in fungal or archaeal abundances were found. Using pooled data, there were no significant differences in the Firmicutes:Bacteroidetes ratio; however, a significant decrease in the relative abundance of Actinobacteria was observed. Additionally, results revealed an increased relative abundance of the genera Roseburia, Clostridium, Dialister and Lachnospira. A decreased relative abundance of Parabacteroides and Bifidobacterium was also observed. When differentiating results by the impact of almond processing, it was found that chopped almonds increased the relative abundance of Lachnospira, Roseburia and Oscillospira, whole roasted almonds increased the relative abundance of Lachnospira and whole raw almonds increased the relative abundance of Dialister, with no difference between almond butter and controls.
Using both adult and children subjects, Burns et al.(Reference Burns, Zitt and Rowe32) used almonds (an option of whole raw almonds or almond butter equivalent) over two 3-week diet periods, with adults consuming 43 g/d and children 14 g/d. The present study found no significant change in microbial diversity or significant shifts at the phylum level. However, significant differences in the prevalence of unclassified bacterial signatures at the genus and species level were reported.
Hazelnuts
Gargari et al.(Reference Gargari, Deon and Taverniti30) investigated the effects of nut consumption on the microbiota over an 8-week period in both children and adults, using 0·43 g of hazelnuts per d per kg of body weight, up to a maximum of 30 g of hazelnuts per d. This was the only study to find no statistically significant differences in microbial diversity or composition of the gut microbiota.
Mixed intervention (almonds + pistachios)
Using a randomised, cross-over design, Ukhanova et al. (Reference Ukhanova, Wang and Baer34) investigated the effects of both almond and pistachio consumption on the gut microbiota, utilising 0, 43 or 85 g/d (equal to 0, 1·5 or 3 serves) for three 18-d feeding periods. While no significant changes in α- or β-diversities were reported, the study did report the pistachio-consuming group to have a greater mean UniFrac distance (unspecified if weighted or unweighted metric used) compared with the almond-consuming group. This suggests that pistachios may have had a greater impact on the overall gut microbiota composition compared with almonds. While many OTUs were affected by nut consumption, only two were significantly decreased: one unspecified OTU closest to the Firmicutes bacterium DJF VP44 and the other unspecified OTU closest to Clostridium sp. ASF 356. Nut consumption did not significantly affect the proportion of the most dominant OTUs. As the present study also sequenced fungal DNA, it was found that while no fungal OTU was significantly increased, various fungal OTUs decreased in proportion (P < 0·01) with nut consumption.
Discussion
To our knowledge, this is the first study to systematically explore the overall microbiota-related changes related to nut consumption. This review establishes that while the evidence base is small, intake of nuts in the diet can exert a modulatory effect on the gut microbiota; however, the exact effects were inconclusive across studies.
Microbial diversity has been shown to be a key predictor of gut health, as greater diversity often equates to greater resilience of the community to recover from or adjust to disturbances(Reference Heiman and Greenway41). On the contrary, a loss of species diversity and an imbalance in the gut’s microbial community, or ‘dysbiosis’, has commonly been found in several disease states(Reference Turnbaugh, Hamady and Yatsunenko9,Reference Frank, St Amand and Feldman42) . The present systematic review found that nut intake may have a modest effect on the diversity of the gut microbiota, with two(Reference Garcia-Mantrana, Calatayud and Romo-Vaquero29,Reference Dhillon, Li and Ortiz33) of the eight total studies reporting a significant change in α-diversity (one showing a decrease in diversity(Reference Garcia-Mantrana, Calatayud and Romo-Vaquero29)), and four(Reference Holscher, Guetterman and Swanson27–Reference Garcia-Mantrana, Calatayud and Romo-Vaquero29,Reference Dhillon, Li and Ortiz33) reporting a significant change in β-diversity. The lack of effect on α-diversity found in this review is similar to the results of another systematic review on dietary fibre(Reference So, Whelan and Rossi14), where short-term feeding studies of dietary fibre in various forms did not increase α-diversity. Furthermore, these findings are also comparable to other dietary intervention studies using whole grains(Reference Vuholm, Nielsen and Iversen43,Reference Vanegas, Meydani and Barnett44) . Interestingly, one study(Reference Garcia-Mantrana, Calatayud and Romo-Vaquero29) included in this review observed a decrease in α-diversity following a 3-d walnut feeding intervention, a result comparable to findings from a short-term dietary intervention advising increased fibre intake(Reference Klimenko, Tyakht and Popenko45). The researchers suggested this temporary reduction in diversity may be due to the rapid dietary change causing disruption to the microbial composition, termed the ‘shock effect’. However, these results are in contrast to previous observational studies that explore the relationship between overall dietary pattern and gut microbiota diversity. For example, adherence to the Mediterranean diet has been associated with increased gastrointestinal bacterial α-diversity(Reference Garcia-Mantrana, Selma-Royo and Alcantara46,Reference De Filippis, Pellegrini and Vannini47) , a finding of particular relevance given that nuts are a key component of this dietary pattern. Given that it has been shown that diet-induced shifts in the gut microbiota occur within 3–4 d after a change in diet(Reference Walker, Ince and Duncan48), the discrepancies observed between the short-term interventional trials included in this review and previous observational studies which assess habitual diets are unlikely due to the study duration alone. Rather, the discrepancies might also be explained by the synergistic effect of an overall healthy dietary pattern, which appears to be a greater determinant of gastrointestinal health than just foods or nutrients in isolation.
Given the variation in the composition of different nut types, consideration of study results by nut type was warranted. Different nut types offer unique nutritional profiles, with altered ratios of fibre, proteins, fat, micronutrients and other bioactive molecules including polyphenols. Of all the nuts investigated in the review, walnuts appeared to more frequently explain variations in the overall gut microbial composition between study participants, with all three of the walnut studies reporting a significant shift in relevant β-diversity metrics. While it is difficult to draw conclusions from this due to the small sample size, the unique nutritional composition of walnuts as a potential modulator of the gut microbiota should be considered and further explored in larger interventional trials. For example, walnuts have a particularly high ellagitannin (a type of polyphenol, the basic structure being ellagic acid) content compared with other nuts(Reference Abe, Lajolo and Genovese49). It has been shown that gastrointestinal bacteria can metabolise ellagic acid to produce urolithins, which can have beneficial vascular and anti-inflammatory effects(Reference Larrosa, Garcia-Conesa and Espin50,Reference Papoutsi, Kassi and Chinou51) , although studies supporting the role of ellagitannins as a prebiotic are inconclusive(Reference Li, Summanen and Komoriya52,Reference Puupponen-Pimiä, Seppänen-Laakso and Kankainen53) . Further, it has been shown that different UMs can affect the metabolism and bioactivity of polyphenols, as demonstrated by Garcia-Mantrana et al.(Reference Garcia-Mantrana, Calatayud and Romo-Vaquero29). Walnuts are also particularly rich in the n-3 essential fatty acid α-linolenic acid(Reference Maguire, O’Sullivan and Galvin54). Preliminary research on n-3 supplementation in humans has found these fatty acids to exert significant effects on gut microbial composition, with the authors concluding n-3 PUFA may be classified as prebiotics(Reference Costantini, Molinari and Farinon26). It should be acknowledged that much of this research was performed using EPA and DHA, which may be synthesised, albeit often inefficiently, from α-linolenic acid (Reference Burdge and Calder55). As a result, the extent to which α-linolenic acid acts as a possible modulator of the gut microbiota remains still unclear.
In the present review, alterations in the gut microbiota profile were reported across a range of different phylogenic categories, although a lack of consistency across studies was observed. Nevertheless, some interesting patterns were observed relating to Bifidobacterium spp. and the SCFA butyrate, which warrants further discussion. Members of the genus Bifidobacterium are normal inhabitants of the human gastrointestinal tract and have demonstrated positive health benefits(Reference O’Callaghan and van Sinderen56), largely due to their production of acetate, which has anti-inflammatory effects and is used by cross-feeding species as a co-substrate for the production of butyrate(Reference Belenguer, Duncan and Calder57). Additionally, research has shown that the loss of Bifidobacterium species in the gastrointestinal tract has been associated with poor bowel outcomes(Reference Grimm, Westermann and Riedel58). Given that previous research has found an increased abundance of Bifidobacterium with increased fibre(Reference So, Whelan and Rossi14) and polyphenol(Reference Jin, Touyama and Hisada59,Reference Vendrame, Guglielmetti and Riso60) intake, the addition of nuts to the diet may have been expected to produce a bifidogenic effect; however, this effect was inconsistent. Rather, two studies(Reference Holscher, Guetterman and Swanson27,Reference Holscher, Taylor and Swanson31) reported a decreased abundance of Bifidobacterium, one(Reference Garcia-Mantrana, Calatayud and Romo-Vaquero29) reported a significant increase in Bifidobacterium for UM-B (when stratified by UM), and one(Reference Bamberger, Rossmeier and Lechner28) found an increase in one OTU from the Bifidobacterium genus. The possible detrimental impact of walnut consumption on Bifidobacterium may be explained by inhibition of Bifidobacterium animalis spp. lactis in the presence of ellagic acid. However, this finding is not universal to the Bifidobacterium genus and appears to be species-specific(Reference Bialonska, Kasimsetty and Schrader61). Further, this hypothesis would not explain the findings of Holscher et al.(Reference Holscher, Taylor and Swanson31), which observed a decreased abundance of Bifidobacterium following almond consumption. One significant gap in the evidence base which may help explain these findings is the use of more sophisticated multi-omic methods (e.g. shotgun sequencing and metabolomics), a crucial step in explaining both the functional potential of the microbiota and the types of metabolites these microbes can produce.
Findings of the present review are in contrast to the results of a recent review by So et al. (Reference So, Whelan and Rossi14), which found that prebiotic fibre supplementation consumption increased Bifidobacterium species. Interestingly, subgroup analysis which separated the effects of isolated fibres from whole foods found that whole-food intervention of mostly grains and cereals had no effect on Bifidobacterium abundances(Reference So, Whelan and Rossi14). However, the impact of whole foods v. isolated food components on Bifidobacterium populations is not consistent across studies. Mandalari et al.(Reference Mandalari, Nueno-Palop and Bisignano62) highlighted the likely importance of using whole-food interventions over isolated fibres, demonstrating that when almonds were consumed with all constituent parts present (in comparison with almonds with the lipid content removed), a significant increase in the abundance of Bifidobacterium was observed. Taken together, these results suggest a lack of consistency for the effects of whole foods on the gut microbiota, highlighting the need for further research exploring the comparative effect of whole foods and food constituents.
The SCFA butyrate is mainly produced by members of the Firmicutes family including Roseburia spp. Lachnospira spp. and Faecalibacterium prausnitzii, which belong to the clostridium clusters XIVa and IV(Reference Louis and Flint63). Butyrate is a major source of energy for the colonic epithelial cells and has many well-documented health effects, including the inhibition of colonic carcinogenesis and acting as an anti-inflammatory agent(Reference Canani, Costanzo and Leone64). The present review found two studies(Reference Holscher, Guetterman and Swanson27,Reference Holscher, Taylor and Swanson31) to report a significantly increased abundance of Clostridium and Roseburia, another two studies(Reference Holscher, Taylor and Swanson31,Reference Dhillon, Li and Ortiz33) to report a significantly increased abundance of Lachnospira and one study(Reference Ukhanova, Wang and Baer34) to report a significant increase in an OTU closest to Clostridium sp. ASF-356. Studies in vitro have found that finely ground almonds increase concentrations of butyrate(Reference Mandalari, Nueno-Palop and Bisignano62) and that Roseburia may be a major contributor to the butyrate-producing capacity of the gastrointestinal tract(Reference Pryde, Duncan and Hold65). As an increased abundance of specific butyrate-producing species was observed in the included trials listed above, it may be inferred that this potentially resulted in a butyrogenic effect, although further research utilising metabolomic analysis is required to confirm this. In contrast to these findings, one study(Reference Holscher, Guetterman and Swanson27) included in the review did report finding no difference in the predicted number of bacterial butanoate metabolism genes following walnut consumption, potentially explained by a proportionate decrease in other butyrate-producing species during the feeding period. Indeed, considering changes in the microbiota as a whole may be a better indicator of the overall metabolomic capacity of the microbiota, rather than focusing on the isolated microbial shift in individual taxa.
In addition to bacteria, the present review also summarised the overall effects of nut intake on fungal and archaeal populations, providing a more comprehensive picture of the human gut microbiota. While it has previously been reported that diet-induced shifts can modulate the populations of these microbes(Reference Hoffmann, Dollive and Grunberg66), the review found no significant changes to the diversity or the relative abundance of archaea, with only one study(Reference Ukhanova, Wang and Baer34) to report a decrease in various fungal OTUs following nut intake. The small sample size of the review may account for the differences observed in these results, as only one study sequenced fungal DNA(Reference Ukhanova, Wang and Baer34) and two studies sequenced both fungal and archaeal DNA(Reference Holscher, Guetterman and Swanson27,Reference Holscher, Taylor and Swanson31) . Further research in this area is needed in order to form a more comprehensive understanding of fungal and archaeal activity with nut intake.
While exploring shifts in the microbiota according to nut type was the main focus of the review, other commonalities between studies were also investigated to identify patterns in results. One study characteristic of interest was the feeding method utilised. For example, three(Reference Holscher, Guetterman and Swanson27,Reference Holscher, Taylor and Swanson31,Reference Ukhanova, Wang and Baer34) of the eight studies were classified as complete feeding trials, whereby all food provided to participants was controlled (as opposed to free-living trials where nuts were often administered alongside general dietary recommendations). Interestingly, while shifts in microbial diversity were negligible (only one of the three studies reported a change in β-diversity), shifts in bacterial composition were notable. In particular, two studies(Reference Holscher, Guetterman and Swanson27,Reference Holscher, Taylor and Swanson31) reported an increase in Clostridium, Roseburia and Dialister, all of which are known to be capable of producing SCFA(Reference Guo, Zhang and Ma67,Reference Koh, De Vadder and Kovatcheva-Datchary68) . In both studies, a run-in period of 9 d was used to allow for the adaptation of the participant’s gut microbiota following the change in habitual diet, increasing the likelihood of these changes being attributable to nut intake. While the small sample size limits the broader interpretation of these findings, these results highlight the importance of controlling for participants’ background diet as a significant variable.
This review followed a robust methodology, including prospective registration of the review protocol and duplicate screening and inclusion of articles. Additionally, the review only included trials employing next-generation sequencing technology, to ensure the most comprehensive analysis of the microbiota reflecting current analytical techniques, and a means of promoting homogeneity between results. The implementation of this exclusion criteria did mean that the evidence base for analysis was smaller, as trials utilising other microbial determination techniques were excluded (such as bacterial culturing and fluorescence in situ hybridisation). As a limitation, the systematic search was restricted to scientific databases, which may have meant that unpublished studies were not detected. Further, the present review did not assess the metabolomic capacity of the gut microbiota including the measurement of SCFA, meaning interpretation of results is limited due to the challenge of connecting microbial shifts to metabolic pathway activity and health outcomes(Reference Bunesova, Vlkova and Rada69). The high level of heterogeneity between studies should also be acknowledged, including the collection and processing of biological samples, sequencing platform and computational pipelines used. Also, papers differed in their statistical analysis and reporting of gut microbial data, including the presentation of findings for different diversity metrics or only at certain taxonomic levels. Well-designed interventional trials with larger sample sizes, reproducible methods and reporting of results are still needed to exemplify the exact effect of nuts on the gut microbiota.
Despite the small number of studies in this review, a large variation in results was observed. This may be due to the intrinsically complex nature of microbiome research, with marked differences in the inter-individual variation of subject’s gut microbiota previously observed(Reference Walker, Ince and Duncan48). Despite strategies to decrease this variation such as the use of cross-over study designs and excluding participants with gastrointestinal disease or those on recent antibiotic therapy, large variations in the composition and function of individual’s microbiota remain. Previous research has indicated that an individual’s response to any dietary intervention is largely dependent upon their established gut microbiota(Reference Martinez, Lattimer and Hubach70). Given these findings, and the knowledge that a patient’s habitual diet and food diversity is a major determinant of their gut microbiota(Reference De Filippo, Cavalieri and Di Paola71), a more rigorous screening and selection process including pre-intervention dietary analysis may be warranted in an attempt to reduce the inter-microbial differences of individual’s microbiota at baseline. In addition, several limitations in the design of several included studies may have impacted results – for example, a lack of reporting of baseline nut consumption. Therefore, robust study designs which take into account an individual’s baseline dietary intake, as well as considering inter-individual gut microbiota differences are required.
Conclusion
This review suggests that nut intake has the potential to yield a modest effect on the gut microbiota, although the exact effects are inconsistent across studies. Compared with other nut types, walnuts appeared to be the strongest driver of overall gut microbial composition (β-diversity), which may be explained by their unique nutritional composition. Walnuts are rich in polyphenols and n-3 fatty acids, both of which have been found to have prebiotic properties. Overall, it appears gut microbial composition is more affected by nut intake during short-term feeding trials than overall microbial diversity, although the small sample size of this review limits interpretation. Further, whether these compositional changes translate into tangible health outcomes remains to be investigated, ideally with the use of more sophisticated multi-omic methods, such as metagenomic and metabolomic technology to evaluate the overall functional capacity of the microbiota. Inconsistencies associated with trial design and methodology, as well as inter-individual microbiota variances and the vast number of potential confounding variables present in this field of research further complicate interpretation of the results. Future trials aiming to explore the influence of nut intake on the gut microbiota must carefully consider study design, ideally incorporating complete feeding methods with a single food or nutrient modification. Additionally, an assessment of habitual dietary pattern and baseline microbiota composition is recommended in order to minimise the inter-individual composition of the gut microbiota.
Acknowledgements
No financial support for this research was received.
E. F., E. P. N. designed the study; E. F., E. P. N. conducted study screening, application of eligibility criteria and data extraction, with guidance from K. L. and J. S.; E. F. drafted the manuscript; E. F., E. P. N. and K. L., J. S. provided critical review of the manuscript.
E. F., K. L. and J. S. declare they have no competing interests. E. P. N. has previously received funding or hold current grants from Nuts for Life, the International Nut and Dried Fruit Council, and the California Walnut Commission.
Supplementary material
For supplementary materials referred to in this article, please visit https://doi.org/10.1017/S0007114520002925






