Inflammatory bowel diseases (IBD), which include ulcerative colitis and Crohn's disease, are currently thought to be caused by the influence of several genes in combination with complex environmental interactions(Reference Noble, Nimmo, Gaya, Russell and Satsangi1). IBD is a chronic condition with no known cure, requiring a lifetime of care for patients. A large number of compounds, including some food components, can either alleviate or worsen the condition of IBD patients(Reference Goh and O'Morain2).
The biological processes associated with IBD susceptibility are dysregulated immunoregulation, mucosal barrier integrity and microbial recognition, clearance and/or homeostasis(Reference Sartor3). There is evidence that changes in the epithelial barrier can cause a continuous stimulation of immune cells in IBD patients(Reference Langmann, Moehle, Mauerer, Scharl, Liebisch, Zahn, Stremmel and Schmitz4). The multidrug resistance 1 (Mdr1) gene encodes the P-glycoprotein (170 kDa; Abcb1 transporter), which is expressed on epithelial cell surfaces and lymphocytes(Reference Panwala, Jones and Viney5). Mdr1a − / − mice are deficient in these transporters and have an intact immune system, but spontaneously develop colitis triggered by the intestinal bacterial flora(Reference Maggio-Price, Shows, Waggie, Burich, Zeng, Escobar, Morrissey and Viney6). In the absence of the mdr1 pump (Abcb1 transporters), bacterial products and toxins become sequestered within the epithelial cells, causing damage to the intestinal barrier and subsequently initiating the inflammatory process in mdr1a − / − mice(Reference Panwala, Jones and Viney5, Reference Maggio-Price, Shows, Waggie, Burich, Zeng, Escobar, Morrissey and Viney6).
Polyphenol compounds are characterised by one or more hydroxyl groups on one or more aromatic rings and are found naturally in fruits, vegetables, grains, roots and tea. In vitro and in vivo studies have shown that different polyphenol compounds can have anti-allergenic, anti-inflammatory, anti-cancer and/or antioxidant activities(Reference Middleton7).
Curcumin is a polyphenol and the major constituent of turmeric powder, a spice and colouring agent extracted from the root of Curcuma longa and widely used in Asian food. It has been shown that curcumin can inhibit the expression of intercellular adhesion molecule 1 (Icam1) and IL-8 (Il8) genes, and cytokine-mediated Nfkb activation in intestinal epithelial cells(Reference Jobin, Bradham, Russo, Juma, Narula, Brenner and Sartor8). Salh et al. (Reference Salh, Assi, Templeman, Parhar, Owen, Gomez-Munoz and Jacobson9) showed that curcumin reduced the activation of Nfkb and the activity of p38 mitogen-activated protein kinase (Mapk) in the colon of mice with chemically induced colitis. Sivalingam et al. (Reference Sivalingam, Hanumantharaya, Faith, Basivireddy, Balasubramanian and Jacob10) showed that feeding curcumin to rats ameliorated chemically induced oxidative stress in the small intestine. The antioxidant activity of curcumin in liver cells was reported by Sharma et al. (Reference Sharma, Gescher and Steward11).
Rutin (quercetin-3-rutinoside) is a polyphenolic flavonoid found in buckwheat seeds, citrus fruits and tea(Reference Kampkotter, Nkwonkam, Zurawski, Timpel, Chovolou, Watjen and Kahl12). Rutin is rapidly deglycosylated by microbial glycosidases in the large intestine, liberating quercetin in the colon(Reference Kim, Kong, Choi, Yang, Kim, Lim, Neckers and Jung13). Rutin's anti-inflammatory effect has been reported to act through: (1) suppression of pro-inflammatory cytokine production in mice with chemically induced colitis(Reference Kwon, Murakami, Tanaka and Ohigashi14), and (2) inhibition of Tnf and Nfkb activation in rats with chemically induced colitis(Reference Kim, Kong, Choi, Yang, Kim, Lim, Neckers and Jung13). Cruz et al. (Reference Cruz, Galvez, Ocete, Crespo, Sanchez de Medina and Zarzuelo15) reported that rutin attenuated inflammation in rats with chemically induced colitis, by alleviating oxidative stress in the colon.
Studies have shown that curcumin and rutin have anti-inflammatory and antioxidant properties both in vitro and in chemically induced colitis animal models, but their mechanism(s) of action are still not completely understood. The present study was primarily designed to test whether dietary curcumin or rutin can reduce colonic inflammation in mdr1a − / − mice, which spontaneously develop intestinal inflammation similar to Crohn's disease(Reference Panwala, Jones and Viney5, Reference Dommels, Butts, Zhu, Davy, Martell, Hedderly, Barnett, McNabb and Roy16). Second, we used genome-wide microarrays to evaluate gene expression in colon tissue, to gain more insight into the pathways underlying the effect of dietary curcumin or rutin on colonic inflammation.
Materials and methods
Mice, diet and sample collection
Mdr1a − / − mice were purchased from Taconic Farm, Inc. (Germantown, NY, USA). This trial was approved by AgResearch Grasslands Animal Ethics Committee (Palmerston North, New Zealand), according to the Animal Welfare Act 1999.
Thirty six male mdr1a − / − mice, 5–6 weeks old, mean weight 23·6 (sd 0·6) g, were housed individually in cages with pressed wood chips and a plastic tube for environmental enrichment. The room was climate controlled (22–24°C, at approximately 60 % humidity) with a 12 h light–dark cycle. All mice were acclimatised for 1 week before the start of the experiment. Mice were randomly assigned to three treatment groups of twelve animals each. Each treatment group received one of the following diets: (1) control (AIN-76A powdered diet); (2) curcumin (AIN-76A+0·2 % curcumin; C7727; ≥ 94 % curcuminoid content and ≥ 80 % curcumin; Sigma-Aldrich, Inc., St Louis, MO, USA); (3) rutin (AIN-76A+0·1 % rutin; R5143; rutin hydrate ≥ 95 %; Sigma-Aldrich Inc.). The composition of the AIN-76A diet was previously described by Dommels et al. (Reference Dommels, Butts, Zhu, Davy, Martell, Hedderly, Barnett, McNabb and Roy16). Animals received food and water ad libitum.
Mice were weighed three times per week throughout the experiment to determine body-weight changes. Food intake was measured during two periods of 4 d at 15 and 18 weeks of age. Food intake was calculated from the amount of food offered during the 4 d minus the remaining and spilt food at the end of the period. On the day before tissue sampling (21 and 24 weeks of age), to minimise the variation between the last meal and sampling, mice were fasted overnight and food was returned the following morning for 2 h before sampling in staggered groups. Six mice from each dietary group were euthanised via CO2 asphyxiation and cervical dislocation at 21 weeks of age (sample group 1) and the remaining animals at 24 weeks of age (sample group 2). These time points were selected based on previous results(Reference Dommels, Butts, Zhu, Davy, Martell, Hedderly, Barnett, McNabb and Roy16) to verify possible time-dependent changes in the colonic inflammation patterns.
Blood was collected from euthanised mice by cardiac puncture. Plasma was separated from erythrocytes by centrifugation (2000 g; 10 min; 4°C), frozen in liquid N2 and stored at − 80°C. The intestine was removed, cut open lengthwise and flushed with 0·9 % NaCl to remove digesta residues. The proximal half of the colon was cut in two pieces, one for histological evaluation and one for gene expression studies. Samples of the colon tissue for gene expression analysis were frozen in liquid N2 and stored at − 80°C. Samples for histological analysis were stored in formalin solution (10 % neutral buffered) at room temperature.
Histology and serum amyloid A analysis
Formalin-fixed colon samples were embedded in a paraffin block, cut into 5 μm thick sections and stained with haematoxylin and eosin for light microscopic examination. The histological scoring of the inflammation was done with no prior knowledge of the treatments. Scoring of stained sections was based on: (1) inflammatory lesions (mononuclear cell, neutrophil, eosinophil and plasmocyte infiltration, fibrin exudation and lymphangiectasis); (2) tissue destruction (enterocyte loss, ballooning degeneration, oedema and mucosal atrophy); (3) tissue repair (hyperplasia, angiogenesis, granulomas and fibrosis). A score between 0 (no change from normal tissue) and 3 (lesions present in most areas and all layers of the intestinal section including mucosa, muscle and omental fat) was given for each aspect of inflammatory lesions, tissue destruction and tissue repair. The histological injury score (HIS) was calculated as: HIS = ((inflammatory lesions score) × 2)+tissue destruction score+tissue repair score.
The concentration of serum amyloid A (Saa) in the plasma was measured to gain information on systemic inflammation levels and to complement HIS. Plasma (10 μl; diluted 1:5000) was used to assess the Saa concentration using the serum amyloid A (murine) kit (Tridelta Development Limited, Maynooth, County Kildare, Republic of Ireland), as described by the manufacturer.
RNA extraction
Total RNA was extracted from the colon tissues using Trizol (Invitrogen, Carlsbad, CA, USA) as described by the manufacturer, with a subsequent purification step using RNeasy columns (Qiagen, San Diego, CA, USA). RNA was quantified using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) and RNA quality was examined using an RNA 6000 LabChip Kit and a 2100 Bioanalyser (Agilent Technologies, Palo Alto, CA, USA). Only total RNA with a 260:280 nm ratio ≥ 2·0, a 28S:18S peak ratio ≥ 0·8 and an RNA integrity number ≥ 6·5 was used for microarray and quantitative real-time PCR analysis.
Microarray hybridisations
As only curcumin significantly reduced colonic HIS, comparison of the gene expression levels in colon was carried out using total RNA from colon tissue of four mdr1a − / − mice from the control group (high HIS) and four mdr1a − / − mice from the curcumin group (low HIS). A reference design with eight arrays was used for this comparison, where each individual RNA sample was hybridised in the array with the reference RNA, in a total of four biological replicates per treatment. Reference RNA was produced by pooling equal amounts of total RNA extracted from the small intestine, colon, kidney and liver of normal healthy Swiss mice plus RNA extracts from Swiss mouse fetuses.
cDNA and cRNA were synthesised using the Low RNA Input Linear Amplification Kit (Agilent Technologies), following the manufacturer's instructions. Reverse transcription used 500 ng of total RNA and oligo-dT primers. cRNA was synthesised and labelled with cyanine3 (samples) and cyanine5 (reference) dye (Perkin-Elmer/NEN Life Science, Boston, MA, USA) and the dye incorporation evaluated using a Nanodrop ND-1000 spectrophotometer. Samples ranged from 10 to 22 pmol dye/μg RNA. Labelled cRNA were hybridised using an In Situ Hybridization Plus Kit (Agilent Technologies), following the manufacturer's protocols, using 750 ng of experimental and reference cRNA onto 44K mouse oligonucleotide arrays (Agilent Technologies). After hybridisation, slides were washed in solutions I, II and III (Agilent Technologies) and air-dried. Slides were scanned using a GenePix Professional 4200A scanner (Molecular Devices, Sunnyvale, CA, USA) with a photomultiplier tube voltage of 580 V. All slides were individually checked and manually flagged for abnormalities. The microarray data are available as accession GSE10684 in the Gene Expression Omnibus repository at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/geo/info/linking.html).
Quantitative real-time polymerase chain reaction
Six genes were selected from genes differentially expressed in the microarray analysis for quantitative real-time PCR verification of the microarray results. cDNA was synthesised from the same total RNA samples used for the microarray analysis, using the Transcriptor First Strand cDNA Synthesis Kit (Roche Diagnostics, Mannheim, Germany). Reverse transcription used 1·0 μg of total RNA and oligo-dT primers (50 pmol) according to the manufacturer's instructions. The PCR conditions were 95°C for 5 min, then thirty-five cycles at 95°C for 15 s, 63°C for 10 s and 72°C for 15 s. Melting curves were assessed by increasing the temperature by 1°C/s from 65°C to 95°C.
Primers for the selected genes were as follows: cytochrome P450, family 2, subfamily C, polypeptide 40 (Cyp2c40, NM_010004.1 – forward primer: ATAGGATGTGCTCCCTGC; reverse primer: AAGGTCTCTGTCCCACCA); carboxylesterase 2 (Ces2, NM_145603.1 – forward primer: CACGGAACCAACTACATAAC; reverse primer: AGACATAGGGAAGGAAGACA); sulfotransferase 1A member 1 (Sult1a1, NM_133670.1 – forward primer: GGATGTGGTTGTCTCCTATT; reverse primer: GGATGGTTGTGTAGTTAGCC); interferon γ (Ifng, NM_008337.1 – forward primer: AAGTGGCATAGATGTGGAA; reverse primer: GCTTATGTTGTTGCTGATG); CD44 antigen (Cd44, AK045226.1 – forward primer: AACACACCAAGAAGACATCG; reverse primer: CCTGATCTCCAGTAGGCTGT) and neutrophil cytosolic factor 4 (Ncf4). Primers (except Ncf4 (Reference Dommels, Butts, Zhu, Davy, Martell, Hedderly, Barnett, McNabb and Roy16)) were designed using Primer 3.0(Reference Rozen, Skaletsky, Krawetz and Misener17), with available public sequences. PCR conditions for all primers were optimised and amplicons were sequenced to confirm identity. Specificities of all PCR reactions were verified in melting curves analyses and in agarose gel. Data were normalised against the geometrical mean of two reference genes (hypoxanthine guanine phosphoribosyl transferase 1 (Hprt) and calnexin (Canx)(Reference Dommels, Butts, Zhu, Davy, Martell, Hedderly, Barnett, McNabb and Roy16)).
Threshold cycle (Ct) values were obtained in duplicate in independent runs for each sample on a Rotor Gene 3000 (Corbett Research, Sydney, NSW, Australia) using LightCycler 480 SYBR Green I Master (Roche Diagnostics), according to the manufacturer's protocol for 20 μl reactions. PCR efficiencies for all runs and primer sets were calculated for each individual sample using the LinRegPCR program(Reference Ramakers, Ruijter, Deprez and Moorman18). As the PCR efficiencies were similar in all reactions (average 2·03 (sd 0·04)), the 2− ΔΔCT method was used for relative quantification.
Pathway analyses
Data from the microarray experiment, comparing colonic gene expression from mdr1a − / − mice fed control and curcumin diets, were used in the pathway analyses to gain more insight into pathways underlying the beneficial effect of curcumin on colonic inflammation. Pathways were generated using the Ingenuity Pathways Analysis (IPA) program (Ingenuity® Systems, Redwood City, CA, USA; www.ingenuity.com) that enables the identification of biologically relevant networks.
Data from microarray analysis containing probe identifiers (gene accession number), the corresponding change in expression value (fold change) and false discovery rate (shown by q values) were uploaded into the IPA program. Only probes from the microarray dataset that met the cut-off criteria of a 1·5-fold change in expression (up- or down-regulated) and a q value of less than 0·05 were considered for the pathway analysis. Each probe identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base. Biological function and relevant metabolic and signalling pathways were assigned to each gene and the most significant pathways ranked according to the significance of the data.
Statistical analysis
Statistical analysis of body weight, food intake, histology and Saa data was performed using ANOVA, followed by the least significant difference post hoc test (GenStat for Windows version 9; VSN International Ltd, Hemel Hempstead, Herts, UK). For all ANOVA analyses, the data were log-transformed before analysis if necessary, to ensure that the variances for all groups were similar. Gene expression analysis was only performed if significant changes in HIS (P < 0·05) were observed in the colon tissue.
Microarray data were analysed using the Limma package in Bioconductor(Reference Smyth, Gentleman, Carey, Dudoit, Irizarry and Huber19). Image analyses of the extracted foreground and background fluorescence intensity measurements were evaluated to select the best background correction for the experiment, but no background correction was necessary due to homogeneous hybridisation. Data were normalised using a local linear regression analysis (LOESS) to remove the effect of systematic variation in the microarrays. Box plots, density plots and spatial images of the raw and normalised data were examined in order to check the quality of the microarray data, and that no unusual results for any slide were observed. Data were log-transformed before analysis and the mean difference between treatments calculated on this scale, resulting in a log ratio for each probe. MA plots of the microarray data were drawn in order to check that there was no dependence of the log ratio on the intensity for any slide. The significance of the log ratio for each probe was determined by calculating one modified t statistic per probe using an empirical Bayesian approach(Reference Smyth20).
The probability values were then corrected for multiple testing using the Benjamini–Hochberg correction(Reference Benjamini and Hochberg21), and a false discovery rate calculated. Probes that had a false discovery rate of less than 5 % (q < 0·05) were considered to be differentially expressed between the groups fed the control and curcumin diets. Log ratios have been converted to fold change when presenting results, as fold change is a more meaningful variable in biological terms.
Pathways were considered significantly affected by curcumin when the probability value calculated using the right-tailed Fisher's exact test was smaller than 0·01 and at least 10 % of the genes from a particular pathway were differentially expressed in the microarray experiment.
One-way ANOVA (GenStat for Windows version 9; VSN International Ltd) was used for statistical analyses of the quantitative real-time PCR data, and gene expression was considered different when the probability was inferior to 0·05.
Results
Weight gain and food intake
There were no differences (P>0·05) in body weight between the mice fed the different diets at 21 and 24 weeks of age (Table 1). The average body weight of the mice is presented in Fig. 1, showing the average growth curve during the experiment. Food intake was similar (P>0·05) between mice fed all three diets at 15 and 18 weeks of age (Table 1). Some animals died during the experiment; three mice from the group fed the control diet, three mice from the group fed the rutin diet and two mice from the group fed the curcumin diet. These mice showed signs of colonic inflammation but were not included in the analyses because they died before the scheduled sampling times.
Table 1 Body weight (g) and food intake (g) during the experiment of multidrug resistance gene-deficient (mdr1a −/−) mice fed control, curcumin and rutin diets
(Mean values and standard deviations)
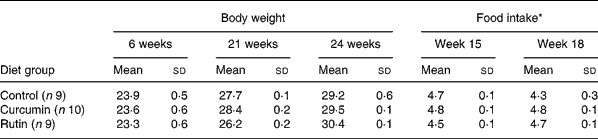
* Food intake was measured during two periods of 4 d at 15 and 18 weeks of age.
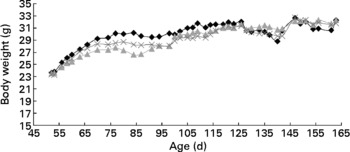
Fig. 1 Body weight of multidrug resistance gene-deficient (mdr1a − / − ) mice fed control (–♦–), rutin (![]() ) and curcumin (
) and curcumin (![]() ) diets as a function of age. Values are means for each time point. No difference was observed in the body weight of mice fed the different diets.
) diets as a function of age. Values are means for each time point. No difference was observed in the body weight of mice fed the different diets.
Histology and serum amyloid A
There were no differences on HIS in the colon (P>0·05) between the two sampling groups (21 and 24 weeks of age) within each dietary treatment (data not shown). For this reason, animals were grouped by diet for further analyses. The average HIS was significantly reduced (P = 0·02) in the colon from mice fed curcumin compared with those fed the control or rutin diets. Table 2 shows that HIS was mainly characterised by cell infiltration (monocytes and neutrophils) and by tissue destruction (enterocyte loss and oedema). Characteristic signs of inflammation were prominent thickening of the mucosal layer, disappearance of crypts, epithelial cell hyperplasia and goblet cell loss in the remaining crypts. Curcumin reduced cell damage and the number of inflammatory cells present in the lamina propria (Fig. 2).
Table 2 Histology injury score (HIS) and principal histological aspects (monocytes, neutrophils, oedema and enterocyte loss) identified in the colon of multidrug resistance gene-deficient (mdr1a −/−) mice
(Mean values)
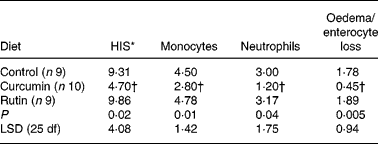
LSD, least significant difference.
* A small part of the HIS includes other histological aspects (data not shown).
† Mean value is significantly different from those of the control and rutin groups (P < 0·05).
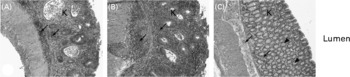
Fig. 2 Multidrug resistance gene-deficient (mdr1a − / − ) colon sections stained with haematoxylin and eosin. Original magnification 100 × . Colon segments of mdr1a − / − mice fed: (A) the control diet, showing signs of inflammation, i.e. a high number of inflammatory cells ( ← ), crypt abscesses and loss of goblet cells in the lamina propria (K); (B) the rutin diet, showing signs of inflammation, i.e. a high number of inflammatory cells ( ← ), crypt abscesses and loss of goblet cells in the lamina propria (K); (C) the curcumin diet, showing reduction of inflammation compared with animals fed the control and rutin diets, i.e. a lower number of inflammatory cells ( ← ) and presence of crypt and goblet cells (◂) in the lamina propria (K).
The concentration of Saa in the plasma normally increases in response to tissue injury and inflammation. Plasma Saa concentration has been suggested as an inflammatory marker to monitor chronic diseases such as arthritis(Reference Cunnane, Grehan, Geoghegan, McCormack, Shields, Whitehead, Bresnihan and Fitzgerald22) and asthma(Reference Ozseker, Buyukozturk, Depboylu, Yilmazbayhan, Karayigit, Gelincik, Genc, Colakoglu, Dal and Issever23). In the present study, the average plasma Saa concentration from mice fed the control diet was 0·424 μg/ml, the curcumin diet 0·135 μg/ml and the rutin diet 0·337 μg/ml. Plasma Saa concentration showed the same trend as the HIS, where curcumin reduced signs of colonic inflammation in mdr1a − / − mice, but was not significantly different (P = 0·27) from the other diets.
Microarray analysis
From the 44 000 oligonucleotide probes in the microarray, 2308 probes were differentially expressed (q < 0·05) in the colon of mice fed the curcumin-enriched diet compared with those fed the control diet. Of these differentially expressed probes, 1319 (57 %) were down-regulated ( − 1·19- to − 9·09-fold) and 989 (43 %) were up-regulated (1·19- to 13·37-fold) in the colon of mdr1a − / − mice fed the curcumin diet compared with those fed the control diet.
Quantitative real-time polymerase chain reaction
To confirm the findings of the microarray analyses, quantitative real-time PCR analyses were carried out on six selected genes from the set of differentially expressed genes. Detoxification genes Ces2, Cyp2c40 and Sult1a1 were up-regulated (P < 0·05) in the colon of the mdr1a − / − mice fed the curcumin diet compared with those fed the control diet. Genes Ifng, Cd44 and Ncf4, which are involved in the immune response process, were down-regulated in the colon of the mice fed the curcumin diet (P < 0·05) compared with the colon of the mice fed the control diet. Similar expression patterns were observed for the six genes tested by quantitative real-time PCR and microarray (Fig. 3), suggesting that the results from the microarrays could confidently be used to identify pathways altered by curcumin.
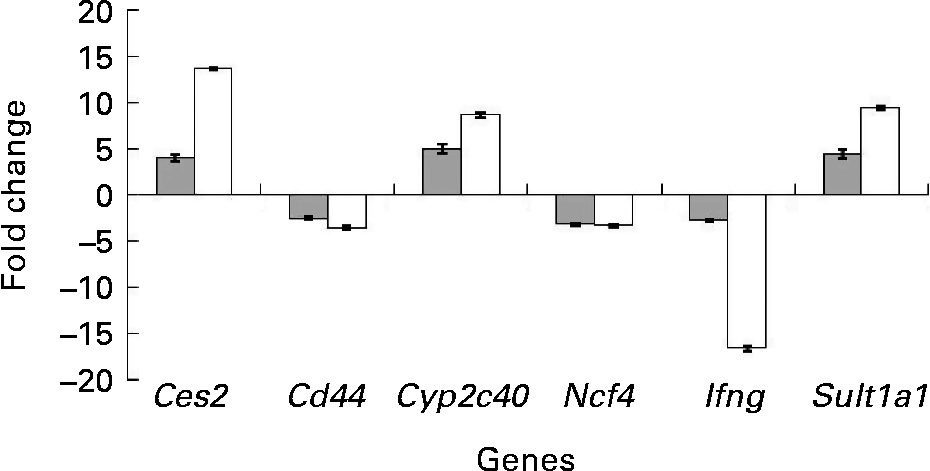
Fig. 3 Change in gene expression of fold change (n 4) in the colon of multidrug resistance gene-deficient (mdr1a − / − ) mice fed the curcumin diet compared with mice fed the control diet for selected genes tested by microarray (![]() ) and quantitative real-time PCR (□). Values are means, with standard deviations represented by vertical bars. Gene expression was significantly different for the six genes in both microarray (q < 0·05) and quantitative real-time PCR (P < 0·05). Quantitative real-time PCR data were normalised for the geometric mean of reference genes. Cd44, CD44 antigen; Ces2, carboxylesterase 2; Cyp2c40, cytochrome P450 family 2 subfamily C polypeptide 40; Ifng, interferon γ; Ncf4, neutrophil cytosolic factor 4; Sult1a1, sulfotransferase family 1A phenol-preferring member 1.
) and quantitative real-time PCR (□). Values are means, with standard deviations represented by vertical bars. Gene expression was significantly different for the six genes in both microarray (q < 0·05) and quantitative real-time PCR (P < 0·05). Quantitative real-time PCR data were normalised for the geometric mean of reference genes. Cd44, CD44 antigen; Ces2, carboxylesterase 2; Cyp2c40, cytochrome P450 family 2 subfamily C polypeptide 40; Ifng, interferon γ; Ncf4, neutrophil cytosolic factor 4; Sult1a1, sulfotransferase family 1A phenol-preferring member 1.
Pathway analyses
The 2308 oligonucleotides probes differentially expressed in the microarray experiment (q < 0·05) corresponded to 830 annotated genes (Ingenuity Pathways Knowledge Base) that met the cut-off (1·5-fold change and q < 0·05). From those, 757 genes were associated with a biological function or disease in the IPA program and used for pathway analysis.
The most significant biological functions affected by dietary curcumin were grouped into three IPA categories: (1) diseases and disorders; (2) molecular and cellular functions; (3) physiological system development and function. The top five functions in these categories and the number of genes differentially expressed in the microarray experiment that were grouped in those functions are shown in Table 3.
Table 3 Significant functions (P<0·0001) affected by dietary curcumin as assessed by Ingenuity Pathways Analysis (IPA)*
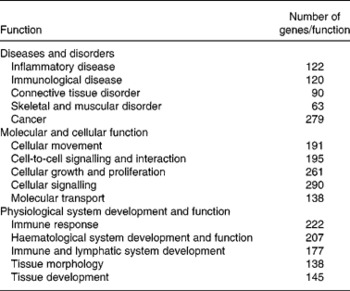
* Functions were grouped into three IPA categories; the Table shows the number of genes differentially expressed in the microarray experiment grouped within a given function.
From the 757 genes associated with a biological function in IPA, 222 genes (29 %) were identified as immune response genes and 145 genes (19 %) were associated with tissue development (Table 3), suggesting an effect of curcumin in immune response and tissue repair. These results support the colonic HIS results, in which mice fed dietary curcumin had colons with reduced cell damage and fewer inflammatory cells in the lamina propria.
The following pathways involved in immune and inflammatory response were affected by dietary curcumin: (1) complement and coagulation cascades; (2) Il10 signalling; (3) lipopolysaccharide/Il1-mediated inhibition of retinoid X receptor (Rxr) function; (4) leucocyte extravasation signalling; (5) Il6 signalling; (6) p38 Mapk signalling; (7) natural killer-cell signalling; (8) Nfkb signalling pathway; (9) positive acute-phase response proteins (P < 0·001 and more than 10 % of the genes belonging to those pathways were differentially expressed). Genes involved in immune and inflammatory responses were generally down-regulated in the colon of the mice fed curcumin compared with those fed the control diet (Table 4), suggesting an anti-inflammatory effect.
Table 4 Immune and inflammatory response genes differentially expressed in the colon of multidrug resistance gene-deficient (mdr1a −/−) mice fed the curcumin diet compared with the colon of mice fed the control diet*
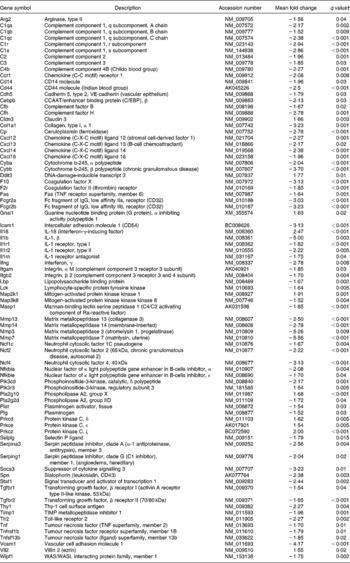
* Genes were grouped into one or more of the following Ingenuity Pathways Analysis pathways: complement and coagulation cascades, Il10 signalling, lipopolysaccharide/Il1-mediated inhibition of retinoid X receptor function, leucocyte extravasation signalling, Il6 signalling, p38 mitogen-activated protein kinase signalling, natural killer-cell signalling, Nfkb signalling and positive acute-phase response proteins.
† Only genes with a false discovery rate < 5 % (q < 0·05) and fold change >1·5 were considered for pathway analysis.
An effect of dietary curcumin was also observed in the following pathways: (1) xenobiotic metabolism signalling; (2) metabolism of xenobiotic by cytochrome P450; (3) cytochrome P450 panel – substrate is a xenobiotic; (4) xenobiotic metabolism; (5) oxidative stress (Table 5). Genes involved in xenobiotic metabolism were up-regulated. Genes involved in the oxidative stress response were generally down-regulated, suggesting an antioxidative effect of the curcumin diet in the colon of the mdr1a − / − mice.
Table 5 Xenobiotic metabolism and oxidative stress genes differentially expressed in the colon of multidrug resistance gene-deficient (mdr1a −/−) mice fed the curcumin diet compared with the colon of mice fed the control diet*
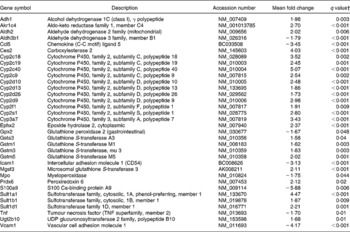
* Genes were grouped into one or more of the following Ingenuity Pathways Analysis pathways: xenobiotic metabolism signalling, metabolism of xenobiotic by cytochrome P450, cytochrome P450 panel – substrate is a xenobiotic, xenobiotic metabolism and oxidative stress.
† Only genes with a false discovery rate < 5 % (q < 0·05) and fold change >1·5 were considered for pathway analysis.
Pathways related to Rxr activity were also affected by curcumin: lipopolysaccharide/Il1-mediated inhibition of Rxr function, pregnane X receptor (Pxr)/Rxr activation, constitutive androstane receptor (CAR)/Rxr activation, vitamin D receptor/Rxr activation and Pparα/Rxrα activation. Genes from those pathways were differentially expressed in the colon of curcumin-fed mice compared with the control mice (Table 6), suggesting that curcumin could affect expression of several genes through those pathways as Rxr regulates the expression of several genes involved in xenobiotic metabolism and immune response. The effect of curcumin on colonic inflammation could indeed be through those key receptors.
Table 6 Genes involved in retinoid X receptor (Rxr) activation pathways differentially expressed in the colon of multidrug resistance gene-deficient (mdr1a −/−) mice fed the curcumin diet compared with the colon of mice fed the control diet*
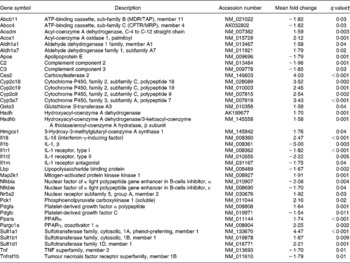
* Genes were grouped into one or more of the following Ingenuity Pathways Analysis pathways: lipopolysaccharide/Il1-mediated inhibition of Rxr function, pregnane X receptor/Rxr activation, constitutive androstane receptor/Rxr activation, vitamin D receptor/Rxr activation and Ppara/Rxrα activation.
† Only genes with a false discovery rate < 5 % (q < 0·05) and fold change >1·5 were considered for pathway analysis.
Two pathways involved in fibrosis development (hepatic fibrosis and hepatic stellate cell activation) were significantly affected by dietary curcumin. Genes from those pathways were down-regulated (Table 7), suggesting that curcumin could inhibit the fibrogenesis process in colon tissue.
Table 7 Genes involved in fibrogenesis differentially expressed in the colon of multidrug resistance gene-deficient (mdr1a −/−) mice fed the curcumin diet compared with the colon of mice fed the control diet*

* Genes were grouped in one or both of the following Ingenuity Pathways Analysis pathways: hepatic fibrosis and hepatic stellate cell activation.
† Only genes with a false discovery rate < 5 % (q < 0·05) and fold change >1·5 were considered for pathway analysis.
Pathways involved in amino acid and fatty acid metabolism were also affected by dietary curcumin; genes involved in those pathways were generally up-regulated (Table 8).
Table 8 Genes linked to amino acid and fatty acid metabolisms differentially expressed in the colon of multidrug resistance gene-deficient (mdr1a −/−) mice fed the curcumin diet compared with the colon of mice fed the control diet*
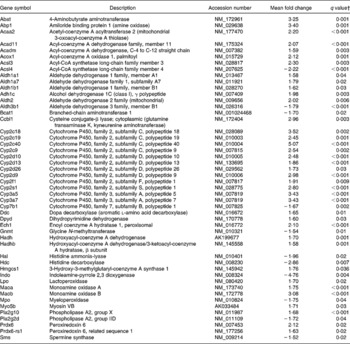
* Genes were grouped in one or more of the following Ingenuity Pathways Analysis pathways: β-alanine metabolism; valine, leucine and isoleucine degradation; histidine metabolism; tryptophan metabolism, phenylalanine metabolism, fatty acid metabolism, linoleic acid metabolism.
† Only genes with a false discovery rate < 5 % (q < 0·05) and fold change >1·5 were considered for pathway analysis.
Discussion
In the present study, mdr1a − / − mice fed a diet containing curcumin had a lower colonic HIS than those fed the control diet. This supports previous studies, which have shown curcumin as a potent anti-inflammatory agent(Reference Salh, Assi, Templeman, Parhar, Owen, Gomez-Munoz and Jacobson9, Reference Sivalingam, Hanumantharaya, Faith, Basivireddy, Balasubramanian and Jacob10). To our knowledge this is the first study using genome-wide gene expression to investigate the effect of curcumin on colonic inflammation in an animal model that spontaneously develops colitis. Microarrays can be used to identify genes and/or pathways that could be relevant to IBD, allowing an unbiased overview of the disease's pathogenesis, and this approach may uncover new targets for further studies and future IBD therapy(Reference Costello, Mah and Häsler24).
Several pathways linked to immune and inflammatory responses were significantly affected by dietary curcumin (Table 4). The expression of the pro-inflammatory cytokines, Il1b, Tnf and interferon γ, previously associated with IBD, was down-regulated in the colon of mice fed dietary curcumin. The expression of several chemokines involved in the leucocyte extravasation signalling pathway was also down-regulated in the colon of those animals. Their down-regulation supports the observed reduction of neutrophil and monocyte infiltration in the lamina propria of the colon from mice fed dietary curcumin (Table 2).
Reyes-Gordillo et al. (Reference Reyes-Gordillo, Segovia, Shibayama, Vergara, Moreno and Muriel25) suggest that curcumin prevents chemically induced acute liver damage by at least two mechanisms: (1) acting as an antioxidant, and (2) inhibiting Nfkb activation and thus reducing the production of pro-inflammatory cytokines. In the present study, genes involved in the oxidative stress and the Nfkb signalling pathways were mostly down-regulated by the curcumin-supplemented diet, and this suggests that curcumin may affect those pathways in the colon.
Genes involved in the p38 Mapk signalling pathway (Fas, Il1b, Il1r1, Il1r2, Il1rn, Pla2g2d, Stat1, Tgfbr1, Tgfbr2, Tnf and Tnfrsf1b) were also down-regulated in the colon of mice fed dietary curcumin compared with the controls. This pathway was previously reported to be inhibited by curcumin in the colon of rats with chemically induced colitis(Reference Camacho-Barquero, Villegas, Sánchez-Calvo, Talero, Sánchez-Fidalgo, Motilva and Alarcón de la Lastra26). Receptors such as Tnf, Il1 receptor, toll-like receptor 2, monocyte differentiation antigen CD14, which regulate the activation of Nfkb and Mapk pathways(Reference Sartor3), were down-regulated in the colon of mice fed dietary curcumin. This suggests that the effects of curcumin in the inflammatory responses in the colon of mdr1a − / − mice may be through Nfkb and Mapk pathways.
Genes involved in pathways of the xenobiotic metabolism were significantly up-regulated in the colon of mice fed dietary curcumin. Detoxification genes, including phase 1 and 2 enzymes such as cytochrome P450 (Cyp enzymes), carboxylesterase (Ces2), glutathione S transferases (Gsta3, Gstm1, Gstm2, Gstm3, Gstm5), UDP-glucuronosyltransferase (Ugt2b10) and sulfotransferases (Sult1a1, Sult1b1, Sult1d1), were up-regulated in colon of mice fed curcumin (low-HIS) compared with those fed the control diet (high-HIS). These results support those observed by Langmann et al. (Reference Langmann, Moehle, Mauerer, Scharl, Liebisch, Zahn, Stremmel and Schmitz4), who showed a down-regulation of detoxification genes in inflamed colon biopsies of IBD patients compared with control patients and our previous results(Reference Dommels, Butts, Zhu, Davy, Martell, Hedderly, Barnett, McNabb and Roy16) where we observed a down-regulation of detoxification genes in the inflamed colonic epithelial cells of mdr1a − / − mice compared with the healthy FVB mice.
Pathways linked to the activation of Rxr were also significant in the pathway analysis. This receptor is the heterodimeric partner that mediates the effects of several nuclear hormone receptors such as Pxr, CAR, vitamin D receptor and Ppara, and regulates the expression of a large number of genes including those involved in xenobiotic metabolism(Reference Waxman27). Rxr homo- or heterodimers have been also suggested to modulate the immune response and T helper cell differentiation(Reference Spilianakis, Lee and Flavell28, Reference Stephensen, Borowsky and Lloyd29). These results suggest that curcumin's effect on the xenobiotic metabolism and immune response could be regulated through those receptors in the colon of mdr1a − / − mice.
Rxr was not differentially expressed in the colon of curcumin-fed mice in the present study. However, this finding does not invalidate the hypothesis that dietary curcumin influences colon inflammation through those receptors. Indeed, previous study showed no marked change in the mRNA transcription levels of Rxr and suggests a post-translational regulation of the Rxr mRNA levels in the acute-phase response in hamster liver(Reference Beigneux, Moser, Shigenaga, Grunfeld and Feingold30).
The transcriptional regulator Pxr was up-regulated (1·38-fold change) in the colon tissue of mice fed the curcumin diet. This supports the results of Kluth et al. (Reference Kluth, Banning, Paur, Blomhoff and Brigelius-Flohe31), which showed that curcumin increases PXR gene expression in human liver carcinoma cells. The constitutive androstane receptor (CAR or Nr1i3) gene was not differentially expressed here. This supports results obtained by Langmann et al. (Reference Langmann, Moehle, Mauerer, Scharl, Liebisch, Zahn, Stremmel and Schmitz4), who showed no evidence of an altered CAR gene expression but a reduced amount of Pxr transcripts in colonic tissues from IBD patients. Beigneux et al. (Reference Beigneux, Moser, Shigenaga, Grunfeld and Feingold32) suggest that CAR and Pxr response elements are quite similar and these two nuclear hormone receptors can compensate for each other's effects on their target genes. This may explain why the CAR/Rxr activation pathway was significantly affected by curcumin, but the CAR gene was not.
The Ppara gene was up-regulated (1·74-fold change) in the colon of mice fed dietary curcumin compared with mice fed the control diet. This suggests that Ppara could have an important modulatory role in colonic inflammation. Bunger et al. (Reference Bunger, van den Bosch, van der Meijde, Kersten, Hooiveld and Muller33) have reported Ppara as an important transcriptional regulator involved in regulation of the immune system in the small intestine. Waxman(Reference Waxman27), reviewing the role of CAR, Pxr and Ppara receptors, suggests that they modulate genes involved in xenobiotic metabolism in response to dietary or hormonal stimuli in the liver. In the present study, pathways linked to those receptors were significantly affected by curcumin, suggesting that the observed regulation of the xenobiotic and immune response in the colon of mice fed dietary curcumin might be related to the up-regulation of Pxr and Ppara and activation of Rxr.
Two pathways involved in fibrosis development (hepatic fibrosis and hepatic stellate cell activation) were down-regulated by dietary curcumin. No signs of fibrosis were observed in the histological analysis of colons in the present study, but a series of genes involved in these pathways were down-regulated, suggesting that curcumin could reduce fibrogenesis in the colon. Burke et al. (Reference Burke, Mulsow, O'Keane, Docherty, Watson and O'Connell34) reviewed fibrogenesis in Crohn's disease and reported studies in which insulin-like growth factor 1, tissue inhibitor of metalloproteinase 1, collagen type III and transforming growth factor β receptors genes were up-regulated in fibroblasts and myofibroblasts in stricture of Crohn's disease patients. These and other genes involved in fibrosis development were down-regulated in the colon tissue of mice fed dietary curcumin compared with those fed the control diet (Table 7), suggesting that curcumin could indeed affect this process.
This genome-wide gene expression analysis of the colon from mdr1a − / − mice shows that dietary curcumin affects the expression of genes involved in several pathways of the xenobiotic metabolism, immune and inflammatory responses and fibrosis development. These results support Jagetia & Aggarwal(Reference Jagetia and Aggarwal35), who suggest that curcumin is a pluripotent pharmacological agent acting through multiple molecular pathways. The global gene expression analyses have limitations, as some key points in the pathways can be regulated at the protein level. However, it does allow an unbiased overview of pathways affected by curcumin, showing the importance of this kind of study to understand and identify nutrient and gene interactions.
In the present study, dietary rutin did not reduce HIS in the colon of the mdr1a − / − mice; this could suggest that the lack of effect was due to the low dose used (0·1 % of rutin in the diet) comparing with results reported by Kim et al. (Reference Kim, Kong, Choi, Yang, Kim, Lim, Neckers and Jung13), in which oral administration of 10 mg/kg had an anti-inflammatory effect in rats with trinitrobenzene sulfonic acid-induced colitis. However, Kwon et al. (Reference Kwon, Murakami, Tanaka and Ohigashi14) used 0·01 and 0·1 % of rutin in the diet and also reported an improvement of inflammation in mice with chemically induced colitis. Rutin is a glycoside form of quercetin that is deglycosylated by microbial glycosidases, liberating quercetin in the colon. Kim et al. (Reference Kim, Kong, Choi, Yang, Kim, Lim, Neckers and Jung13) suggest that quercetin is the active component of the rutin-mediated amelioration of inflammation in rats with chemically induced colitis. Ofer et al. (Reference Ofer, Wolffram, Koggel, Spahn-Langguth and Langguth36), reviewing the effect of quercetin and others polyphenols on the transport of xenobiotics, showed that quercetin can interfere with the secretory transport process by interacting with P-glycoprotein (Abcb1 transporters). This mechanism could explain why 0·1 % of rutin (glycoside quercetin) in the diet reduced intestinal inflammation in the chemically induced colitis animal model(Reference Kwon, Murakami, Tanaka and Ohigashi14), but did not attenuate signs of colitis in mdr1a − / − mice. These mice are deficient in Abcb1 transporters and, therefore, quercetin (from rutin) could not interfere with secretory transport of xenobiotics and toxins.
Conclusions
The present study showed that curcumin, but not rutin, reduced signs of colonic inflammation in mdr1a − / − mice. The genome-wide microarray and pathway analyses allowed an unbiased overview of pathways affected by curcumin, showing the importance of this kind of study in understanding and identifying nutrient and gene interactions. The results suggest that curcumin acts through multiple molecular pathways to ameliorate inflammation. Curcumin might alleviate colonic inflammation via an up-regulation of xenobiotic metabolism and a down-regulation of immune response pathways mediated by Pxr and Ppara activation of Rxr. Curcumin also down-regulates genes involved in oxidative stress and fibrogenesis pathways. However, further studies are needed to elucidate the mechanisms involved in the cross-talk between the pathways identified here. These results show the potential of genome-wide microarray and pathway analyses to study the effect of dietary supplements or functional foods that could prevent or reduce colonic inflammation.
Acknowledgements
The present study is part of Nutrigenomics New Zealand, a collaboration between AgResearch, Crop & Food Research, HortResearch and The University of Auckland, funded by the New Zealand Foundation for Research, Science and Technology. The authors acknowledge the important contribution of: Janice Rhodes and Hannah Smith (Crop & Food Research) during the animal trial; Kim Oden (AgResearch) for Saa analyses; Lee Deighton (AgResearch) for microarray scanning and overlay; Julian Heyes and Juliet Sutherland (Crop & Food Research) and Bruce Sinclair (AgResearch) for valuable discussions and manuscript reviewing; Pieter Demmers (Crop & Food Research) for microarray submission in the Gene Expression Omnibus.
The authors' contributions were: K. N. for microarray analysis, quantitative real-time PCR and pathway analysis, and manuscript writing; Y. E. M. D. for experimental design, animal experiment, microarray experiment and manuscript reviewing; S. M. and C. B. for animal experiment and manuscript reviewing; N. R., M. P. G. B. and W. C. M. for science leadership, experimental design and manuscript reviewing; S. Z. for histology analysis; Z. A. P. for microarray statistics and manuscript reviewing; D. H. for other statistics and manuscript reviewing. All authors read and approved the final manuscript. K. N. and Y. E. M. D. contributed equally to the present study.
The authors declare that there are no conflicts of interest about the present study.













