1. Introduction
Transposons are important components of the genomes of many organisms. Their activity causes mutations and chromosome breakage – damage that is best studied in model genetic organisms such as Drosophila melanogaster. Recent genetic and molecular analyses have revealed that Drosophila has elaborate mechanisms to repress transposon activity, and that small RNAs play key roles in some of these mechanisms (Josse et al., Reference Josse, Teysset, Toideschini, Sidor, Anxolabéhère and Ronsseray2007; Chambeyron et al., Reference Chambeyron, Popkova, Payen-Groschêne, Brun, Laouini, Pelisson and Bucheton2008; Jensen et al., Reference Jensen, Stuart, Goodpaster, Goodman and Simmons2008; Brennecke et al., Reference Brennecke, Aravin, Stark, Dus, Kellis, Sachidanandam and Hannon2007, Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008; Klattenhoff et al., Reference Klattenhoff, Xi, Li, Lee, Xu, Khurana, Zhang, Schutz, Koppetsch, Nowosielska, Seitz, Zamore, Weng and Theurkauf2009; Li et al., Reference Li, Vagin, Lee, Xu, Ma, Xi, Seitz, Horwich, Syrzycka, Honda, Kittler, Zapp, Klattenhoff, Schulz, Theurkauf, Weng and Zamore2009; Tushir et al., Reference Tushir, Zamore and Zhang2009). The RNAs that interact with the Piwi class of proteins, called Piwi-interacting RNAs (piRNAs), appear to be especially important.
One of the major loci for the production of piRNAs is situated in the Telomere Associated Sequences (TAS) at the left end of the X chromosome. A transposon inserted in this locus generates both sense and antisense piRNAs (Brennecke et al., Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008). The antisense piRNAs are of particular significance because they can be targeted to sense mRNAs produced by other copies of the transposon elsewhere in the genome. These mRNAs can then be cleaved into small fragments that become sense piRNAs, which may subsequently be targeted to antisense RNAs transcribed from the telomeric locus to generate more antisense piRNAs. With repetition, this alternating, or ping-pong, cycle is expected to produce a large population of sense and antisense piRNAs (Aravin et al., Reference Aravin, Hannon and Brennecke2007; Gunawardane et al., Reference Gunawardane, Saito, Nishida, Miyoshi, Kawamura, Nagami, Siomi and Siomi2007; Brennecke et al., Reference Brennecke, Aravin, Stark, Dus, Kellis, Sachidanandam and Hannon2007, Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008; Li et al., Reference Li, Vagin, Lee, Xu, Ma, Xi, Seitz, Horwich, Syrzycka, Honda, Kittler, Zapp, Klattenhoff, Schulz, Theurkauf, Weng and Zamore2009) and, concomitantly, to destroy transposon mRNA (Jensen et al., Reference Jensen, Stuart, Goodpaster, Goodman and Simmons2008). It has been speculated that some of the piRNAs may also guide transcription-inhibiting proteins to transposon copies present in the genome (Josse et al., Reference Josse, Teysset, Toideschini, Sidor, Anxolabéhère and Ronsseray2007; Simmons et al., Reference Simmons, Thorp, Buschette, Peterson, Cross and Bjorklund2010). Thus, either by destroying transposon mRNA or by preventing its synthesis, piRNAs can undercut transposon expression and repress transposition.
Both retrotransposons and cut-and-paste transposons can be regulated by the piRNAs generated from ping-pong cycling. As a class, the retrotransposons are more numerous and may have a greater genetic and evolutionary significance. However, one cut-and-paste transposon, Drosophila's P element, provides an unusual opportunity to study the ping-pong model of piRNA amplification because this element can be genetically manipulated in crosses.
P elements appear to have entered the genome of Drosophila melanogaster by horizontal transfer during the 20th century, and have since spread worldwide (Kidwell, Reference Kidwell1983). P transposition is catalysed by a transposase encoded by complete members of the P element family (Karess & Rubin, Reference Karess and Rubin1984); incomplete P elements are unable to make the transposase, but they can be mobilized if the transposase is provided by a complete element somewhere in the genome. P-element movement occurs only in the germ line because transposase synthesis is restricted to that tissue (Laski et al., Reference Laski, Rio and Rubin1986). Within the germ line, P movement is regulated by a maternally transmitted condition called the P cytotype, which depends on the P elements themselves (Engels, Reference Engels1979). Genetic analyses have revealed that this condition can be established by P elements inserted in the TAS of the XL telomere (Ronsseray et al., Reference Ronsseray, Lehmann and Anxolabéhère1991, Reference Ronsseray, Lemaitre and Coen1993, Reference Ronsseray, Lehmann, Nouaud and Anxolabéhère1996, Reference Ronsseray, Marin, Lehmann and Anxolabéhère1998; Marin et al., Reference Marin, Lehmann, Nouaud, Izaabel, Anxolabéhère and Ronsseray2000; Stuart et al., Reference Stuart, Haley, Swedzinski, Lockner, Kocian and Simmons2002; Simmons et al., Reference Simmons, Raymond, Niemi, Stuart and Merriman2004; Niemi et al., Reference Niemi, Raymond, Patrek and Simmons2004; Josse et al., Reference Josse, Teysset, Toideschini, Sidor, Anxolabéhère and Ronsseray2007; Jensen et al., Reference Jensen, Stuart, Goodpaster, Goodman and Simmons2008), and molecular analyses with some of these elements have shown that they produce piRNAs (Brennecke et al., Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008). Cytotype regulation of the P-element family therefore appears to be mediated by maternally transmitted piRNAs (Brennecke et al., Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008; Thorp et al., Reference Thorp, Chapman and Simmons2009). Genetic analyses have also revealed that cytotype regulation anchored in a telomeric P element (TP) can be enhanced by numerous non-TPs scattered about the genome even though the latter have no intrinsic regulatory ability (Simmons et al., Reference Simmons, Niemi, Ryzek, Lamour, Goodman, Kraszkiewicz and Wolff2007). This synergism has been postulated to result from a ping-pong cycle fed by antisense RNAs from the TP and sense mRNAs from the other P elements (Belinco et al., Reference Belinco, DiPrima, Wolff, Thorp, Buschette and Simmons2009).
In the laboratory P elements can be activated by crossing them into strains that lack the P cytotype. These strains have the M cytotype, a condition that permits P-element movement. The M cytotype is characteristic of strains that do not have P elements in their genomes (pure M strains) and of some strains that have them; these latter strains are denoted M′. When strains with potentially active P elements (P strains) are crossed with M cytotype strains, the offspring may exhibit a syndrome of germ-line abnormalities called hybrid dysgenesis (Kidwell et al., Reference Kidwell, Kidwell and Sved1977). This syndrome includes traits such as a high mutation rate, frequent chromosome breakage and sterility. The sterility occurs because the germ-line cells in the gonads are wiped out (Khurana et al., Reference Khurana, Wang, Xu, Koppetsch, Thomson, Nowosielska, Li, Zamore, Weng and Theurkauf2011). This phenomenon, called gonadal dysgenesis (GD), is enhanced by culturing the developing flies at 29 °C. Usually only the offspring from crosses between P males and M females exhibit dysgenesis. The offspring from the reciprocal cross, P females × M males, are not dysgenic because the P cytotype is transmitted maternally (Engels, Reference Engels1979).
In this paper, we address several questions about cytotype regulation of the P-element family. Can this regulation be enhanced by introducing just one additional P element into a genotype that has an X-linked TP, and if so, what kinds of additional P elements can bring about the enhanced regulatory state? Can the additional enhancing element be on any of the major chromosomes? Can the enhanced regulatory state be transmitted to offspring independently of either of the two interacting P elements? Does it persist in subsequent generations when these elements are removed from the genotype? Is it sensitive to cytotype-disrupting mutations? Does the ping-pong cycle of piRNA formation provide an adequate explanation for the synergism between TPs and non-TPs. Can this synergism be explained by other molecular mechanisms?
2. Materials and methods
(i) Drosophila stocks and husbandry
Information on the mutant alleles used in the experiments is available on the Flybase website, in Lindsley & Zimm (Reference Lindsley and Zimm1992), or in references cited in the text. The isolation and initial analysis of the TPs, TP5 and TP6 are described in Stuart et al. (Reference Stuart, Haley, Swedzinski, Lockner, Kocian and Simmons2002). All stocks carrying these elements were marked with a wild-type allele of the yellow body locus (y +) and a mutant allele of the white eye locus (w) – both tightly linked to the XL telomere. Genomic Southern blotting and PCR with P-specific primers established that no other P elements were present in these stocks. TP5 strains that were heterozygous for mutations in the genes aubergine (aub), piwi and Suppressor of variegation 205 [Su(var)205], and correlated strains from which these mutations had been removed, are described in Belinco et al. (Reference Belinco, DiPrima, Wolff, Thorp, Buschette and Simmons2009). Maps of TP5, TP6 and the other P elements used in this study are presented in Fig. 1.

Fig. 1. Structures of P elements used in this study. The 31 bp inverted terminal repeats are represented by arrows. Exons are open boxes and introns are lines connecting the boxes. Missing sequences are indicated by dotted lines. The first or last nucleotides in particular segments of the elements are noted with reference to the nucleotides in the 2907 bp-long canonical complete P element, CP. In the H(hsp/P) transgenes, the P element was truncated at nucleotide 38 and at either nucleotide 2688 (for P=TP5, TP6) or nucleotide 2872 (for P=CP, SP). The P* element is a special case. This frameshifted P coding sequence spans nucleotides 153–2706 minus the introns; the frameshift is due to deletion of nucleotide 279. Recombination mapping established the genetic positions of the H(hsp/P) transgenes used in this study: H(hsp/CP)2 (2–9·2 or 34·8; located 12·8 cM from Sp), H(hsp/TP5)D (2–73·6), H(hsp/TP5)X (1–9·5), H(hsp/TP6)C (3–88·2), H(hsp/P*) (3–0·3).
Experimental cultures were reared in vials on a standard cornmeal-molasses-dried yeast medium at 25 °C unless otherwise specified; stock cultures were maintained in vials or in half-pint milk bottles at 18–21 °C.
(ii) Hobo transgenes with P-element sequences
Stocks carrying hobo transgenes with different terminally truncated (and therefore intrinsically immobile) P elements situated downstream from the hsp70 promoter have been described (Simmons et al., Reference Simmons, Haley, Grimes, Raymond and Fong2002a, Reference Simmons, Haley, Grimes, Raymond and Niemi2002b; Jensen et al., Reference Jensen, Stuart, Goodpaster, Goodman and Simmons2008). These transgenes, denoted in general as H(hsp/P), were inserted on the X chromosome, chromosome 2 or chromosome 3. The insertions were obtained by injecting embryos from an M strain that had been established to be free of P elements by PCR using P-specific primers. This strain has been characterized as ‘E’ in the hobo system of hybrid dysgenesis – i.e. devoid of hobo transposase activity and unable to repress hobo transposition induced by crosses with ‘H’ strains. A plasmid encoding the hobo transposase was co-injected with the H(hsp/P) constructs to obtain the transgene insertions. The P elements within the H(hsp/P) transgenes included SP (0·5 kb long), CP (the complete 2·9 kb P element encoding the P transposase), TP5 (1·8 kb long) and TP6 (1·9 kb long); see Fig. 1. All the transgenic stocks were homozygous for the X-linked markers y (conferring yellow body colour) and w67c23 (conferring white eye colour; hereafter denoted simply as w); however, because the transgenes carried a functional mini-white gene, the flies in these stocks had pigmented eyes.
Another transgenic stock was generated by transforming flies with the construct denoted as H(hsp/P*). In this construct, P* is the sequence from nucleotide 153 (the first one in the initiation codon) to nucleotide 2706 (the third one in the last codon of the transposase gene) in the canonical P element (O’Hare & Rubin, Reference O'Hare and Rubin1983), minus all three introns and nucleotide 279, which causes a frameshift mutation in codon 43; thus, P* is a mutated P coding sequence; see Fig. 1. The H(hsp/P*) construct was created in several steps. Firstly, the frameshifted P coding sequence in the plasmid pAR87 was amplified by PCR using the Pfu DNA polymerase and primers complementary to the ends of the sequence; these primers were augmented with oligonucleotides that contained the recognition sequence for the restriction enzyme BamHI, thereby permitting the P* PCR product to be cloned into the BamHI site of another plasmid. Secondly, a 0·45 kb EcoRI/BamHI fragment containing the hsp70 promoter was cloned between the EcoRI and BamHI sites in the plasmid pMartini. Thirdly, the P* PCR product was cut from its plasmid and inserted into the BamHI site downstream of the hsp70 promoter in the pMartini clone. Finally, the hsp70/P* cassette was excised from pMartini using the restriction enzyme NotI, which recognizes sites on either side of the cassette, and the resulting NotI fragment was inserted into the unique NotI site in the hobo transformation vector pHawN (Blackman et al., Reference Blackman, Koehler, Grimalia and Gelbart1989; Calvi & Gelbart, Reference Calvi and Gelbart1993), which contains the mini-white marker. The H(hsp/P*) construct was then introduced into mutant w Drosophila by germ-line transformation using a plasmid source of the hobo transposase. The single insertion that was obtained, denoted H(hsp/P*)B, was localized to chromosome 3 and made homozygous by inbreeding. Genetic map positions of all the transgenes used in this study are given in the legend to Fig. 1.
(iii) RNA isolation and reverse transcription (RT)–PCR
RNA was isolated from groups of 20 females using TRIZOL (Invitrogen) according to the supplier's instructions. The RNA was reverse transcribed into cDNA using the ThermoScript reverse transcriptase (Invitrogen) and a P-specific primer denoted P2575-u (5′-CAACATCGACGTTTCGCGCTG-3′), directed towards the 5′ end of the P element. After adding the primers P▵0/1-d and P▵2/3-u, the resulting cDNA was amplified by the PCR over 30 cycles using an appropriate temperature profile (see Jensen et al., Reference Jensen, Stuart, Goodpaster, Goodman and Simmons2008), and the products were analysed in a 1% agarose gel by electrophoresis at 70 V.
(iv) Assay for GD
Repression of P-element movement is conveniently assayed by scoring females for the inability to produce eggs. This form of sterility, called gonadal dysgenesis (GD), is due to P-induced destruction of the germ-line cells (Nikki & Chigusa, Reference Nikki and Chigusa1986). To assay for GD, we squashed samples of females between two glass slides and looked for eggs. A solution of green food colouring helped to visualize the eggs extruded from each female. Any female that did not extrude eggs was scored as dysgenic.
The flies to be scored were produced by crossing females of a test genotype to males from either the Harwich w (Kidwell et al., Reference Kidwell, Kidwell and Sved1977) or the Harwich y w P strains, which are both powerful inducers of GD in crosses to females from M strains. The Harwich y w strain was created by introducing the y and w67c23 markers into the Harwich w stock. The test females were initially mass mated at 21 °C. After 3 days, they were aspirated into separate cultures, which were incubated at 29 °C. On day 11, all the offspring were transferred to a holding vial, where they matured for 2 days. As many as 20 females of each segregating genotype were then scored for GD.
(v) Statistical analyses
The frequency of GD was calculated independently for each class of offspring in each vial. Unweighted average frequencies and empirical standard errors (se) among all the vials in a test group were then computed for each class. Averages and se for each group were also computed by pooling the raw data across classes in vials. Statistical differences between averages were evaluated by performing t or z tests.
3. Results
(i) Synergisic repression of P-element activity by combinations of telomeric and non-telomeric transgenic P elements
Although cytotype regulation is anchored in the TPs, it is enhanced by other P elements from M′ strains such as M5 Birmingham (Simmons et al., Reference Simmons, Niemi, Ryzek, Lamour, Goodman, Kraszkiewicz and Wolff2007). These strains contain numerous P elements that collectively have little or no regulatory ability. The stronger regulation that occurs in TP-M′ combinations therefore indicates that the telomeric and M′ elements interact synergistically. A single additional P element might be able to bring about this effect. To investigate this possibility, we tested individual transgenic P elements for interactions with the telomeric elements TP5 and TP6.
The test system involved flies that carried a TP on the X chromosome and a P-containing transgene inserted at a non-telomeric location on an autosome. The transgene was designed to express the P element from either of two promoters – the native P promoter, or the heat-shock-inducible hsp70 promoter, which was situated immediately upstream. However, in the experiments reported here, no heat shock treatments were employed. Each transgene was constructed using a hobo transformation vector (symbolized H) marked with the mini-white gene. Different P elements were inserted behind the hsp70 promoter within the hobo element to create four H(hsp/P) transgenes. RT–PCR analysis with the H(hsp/CP), H(hsp/TP5) and H(hsp/TP6) transgenes has shown that each of them produces P mRNA in the female germ line (Jensen et al., Reference Jensen, Stuart, Goodpaster, Goodman and Simmons2008), which is the physiologically relevant tissue for studies of P-element regulation. However, by themselves, neither these transgenes nor the H(hsp/SP) transgene has any ability to repress GD (Simmons et al., Reference Simmons, Haley, Grimes, Raymond and Niemi2002b; Jensen et al., Reference Jensen, Stuart, Goodpaster, Goodman and Simmons2008). These transgenes were mapped by recombination with dominant markers; none of them proved to be near the telomeres or centromeres of chromosomes 2 or 3.
Interactions between the telomeric and transgenic P elements were assayed by scoring the daughters of TP y+ w/y w; H(hsp/P)/+ females for GD, which was induced by crossing these females to Harwich y w males. In these crosses, we could track the inheritance of the TP and the H(hsp/P) transgene by following the body and eye colour markers. Daughters with wild-type body colour carried the TP (which was tightly linked to the y + allele) and daughters with pigmented eyes carried the H(hsp/P) transgene (which contained the pigment-producing mini-white gene). This design allowed us to determine if synergistic repression of GD involved maternal or zygotic effects of the telomeric and transgenic P elements. In addition, we produced the TP y+ w/y w; H(hsp/P)/+ females for the test matings by performing reciprocal crosses between TP y+ w flies and y w; H(hsp/P) flies (TP y+ w as female in cross A and as male in cross B). This feature allowed us to determine if the parental origin of the TP and the H(hsp/P) transgene mattered.
Table 1 summarizes the results of the tests for synergistic regulation involving the telomeric element TP5. In the absence of any transgene, TP5 repressed GD moderately, but only in the daughters of the tested females from cross A. We observed 84% GD when TP5 was present in these daughters and 87% GD when it was absent. In comparison, we observed 98% GD in both classes of daughters from the tested females from cross B. The results from cross A and cross B are significantly different. Thus, as previously reported (Belinco et al., Reference Belinco, DiPrima, Wolff, Thorp, Buschette and Simmons2009; Thorp et al., 2009), in cross A a telomeric TP5 element moderately represses GD in both the daughters that inherit it and in those that do not.
Table 1. Synergism between the telomeric TP5 element and various H(hsp/P) transgenes assessed in the F2 daughters of TP5 y+ w/y w; H(hsp/P)/ + F1 females from reciprocal crosses between TP5 y+ w and y w; H(hsp/P) strains

a Two factors – the telomeric element TP5 and the H(hsp/P) transgene – segregated in the test crosses, giving rise to four genotypic classes in the F2. The headings indicate which of these two factors were present in the females that were scored for GD.
b The H(hsp/CP)2 and H(hsp/TP5)D transgenes are located on chromosome 2 and the H(hsp/SP)A and H(hsp/TP6)C transgenes are located on chromosome 3.
c Cross A is TP5 y+ w females×y w; H(hsp/P) males, and cross B is TP5 y+ w males×y w; H(hsp/P) females.
d Unweighted average percentage GD±se.
To evaluate the cases in which TP5 was combined with an H(hsp/P) transgene, we note that four classes of daughters segregated in these tests. There were daughters with and without TP5, and with and without the transgene. In each test, the GD frequencies were roughly the same across all four classes. Thus, any ability to repress GD must be established in the TP5 y+ w/y w; H(hsp/P)/ + F1 females and transmitted to their F2 daughters independently of either the TP5 element or the H(hsp/P) transgene – that is, repression involves a strictly maternal effect. On this account, the overall frequencies at the right side of Table 1 adequately summarize the data from each test group. When these frequencies are examined, we see that three transgenes [H(hsp/CP)2, H(hsp/TP5)D and H(hsp/TP6)C] significantly boosted the repression caused by TP5 alone. H(hsp/TP5)D was the most powerful enhancer with this TP, engendering very strong repression in the F2 females from cross A (10% GD) and moderate repression in the females from cross B (82·5% GD). H(hsp/CP)2 was almost as powerful (20% GD in cross A and 84·8% GD in cross B) even though it produces the P transposase, which might be expected to increase rather than decrease the frequency of GD. H(hsp/TP6)C was the least effective enhancer; this transgene brought about strong repression in the F2 females from cross A (52·6% GD), but it had no effect in the F2 females from cross B (98·8% GD).
Although the H(hsp/TP5)D, H(hsp/CP)2 and H(hsp/TP6)C transgenes enhanced the ability of the telomeric TP5 element to repress GD, the H(hsp/SP)A transgene did not (87% GD in cross A and 98·5% GD in cross B). The P element in this last transgene is evidently too small to augment the intrinsic repression ability of the TP5 element. Note that in all the cases where enhanced repression was seen, it was stronger in the F2 females from cross A than in those from cross B, and the F2 females that lacked both the telomeric and transgenic P elements were as effective in repressing GD as the females that had one, the other, or both of these elements – a clear demonstration that enhanced repression involves a maternal effect.
Table 2 summarizes the results of the tests for interaction between the telomeric TP6 element and the four H(hsp/P) transgenes. By itself, TP6 was a moderate repressor in the F2 females from cross A (69% GD), regardless of whether or not they inherited TP6. However, it was not a repressor of GD in the F2 females from cross B (98% GD). In the various transgene combinations, TP6 interacted synergistically with H(hsp/TP5)D, H(hsp/CP)2 and H(hsp/TP6)C to repress GD in the F2. In each test, all four classes of F2 daughters showed approximately equal frequencies of GD. Thus, the regulation created by these interactions involves a maternal effect. In addition, the regulation was consistently stronger in the F2 females from cross A than in those from cross B.
Table 2. Synergism between the telomeric TP6 element and various H(hsp/P) transgenes assessed in the F2 daughters of TP6 y+ w/y w; H(hsp/P)/ + F1 females from reciprocal crosses between TP6 y+ w and y w; H(hsp/P) strains

a Two factors – the telomeric element TP6 and the H(hsp/P) transgene – segregated in the test crosses, giving rise to four genotypic classes in the F2. The headings indicate which of these two factors were present in the females that were scored for GD.
b Cross A is TP6 y +w females×y w; H(hsp/P) males, and cross B is TP6 y +w males×y w; H(hsp/P) females.
c Unweighted average percentage GD±se.
Comparison of the data in Table 2 with those in Table 1 indicates that repression by the interacting TP6-H(hsp/P) combinations paralleled repression by the interacting TP5-H(hsp/P) combinations. H(hsp/TP5)D was the most effective interactor with the telomeric TP6 element, H(hsp/CP)2 was the next most effective interactor and H(hsp/TP6)C was the least effective interactor. Furthermore, as in the tests with the telomeric TP5 element, the H(hsp/SP)A transgene did not interact synergistically with the telomeric TP6 element.
(ii) Synergism between the telomeric TP5 element and a non-telomeric X-linked H(hsp/TP5) transgene
The H(hsp/P) transgenes used in the first tests for synergism with the X-linked TPs were all located on autosomes. To determine if an X-linked H(hsp/P) transgene could enhance repression by one of these TPs, we assayed GD in the daughters of females that were repulsion heterozygotes for TP5 and H(hsp/TP5)X, a TP5-containing transgene located at genetic map position 9·5 on the X chromosome. Preliminary control crosses had established that by itself H(hsp/TP5)X has no ability to repress GD.
The TP5 y+ w/y w H(hsp/TP5)X females for the synergism tests were produced in reciprocal crosses between the TP5 y+ w and y w H(hsp/TP5)X strains (cross A used TP5 y+ w females and cross B used TP5 y+ w males). When test crossed to Harwich y w males, these heterozygous females segregated four classes of offspring. However, owing to linkage, two of the classes – one in which the offspring inherited both TP5 and H(hsp/TP5)X and another in which they inherited neither of these elements – were scarce. The GD data from these two classes were therefore determined with less precision than the data from the other two classes.
The results of this experiment are summarized in Table 3. Control crosses in which the telomeric TP5 element was not combined with the H(hsp/TP5)X transgene showed that, as in the initial analyses, the telomeric TP5 element was a moderate repressor of GD, but only in the flies derived from cross A (87% GD). When the telomeric TP5 element was combined with H(hsp/TP5)X, the repression became much stronger – 43·3–56% GD in the flies derived from cross A. In the flies derived from cross B, little or no repression was observed. The strong repression seen in the flies from cross A was detected in all four genotypic classes that came from the test crosses. Thus, it involved a maternal effect of the telomeric and transgenic TP5 elements acting in the mothers of the flies that were scored. The se associated with the data from the two scare classes are too large to claim that the enhanced repression is statistically significant. However, the errors associated with the other two classes are small enough to justify this claim.
Table 3. Synergism between the telomeric TP5 element and the H(hsp/TP5)X transgene assessed in the F2 daughters of TP5 y+ w/y w H(hsp/TP5)X F1 females from reciprocal crosses between TP5 y+ w and y w H(hsp/TP5)X strains

a Two X-linked factors – the telomeric element TP5 and the H(hsp/TP5)X transgene – segregated in the test crosses, giving rise to four genotypic classes in the F2. The headings indicate which of these two factors were present in the females that were scored for GD. The classes with neither or both of these factors are scarce because TP5 and H(hsp/TP5)X are 9·5 cM apart.
b Cross A is TP5 y +w females×y w H(hsp/TP5)X males, and cross B is TP5 y +w males×y w H(hsp/TP5)X females.
c Unweighted average percentage GD±se.
This experiment shows that a different insertion of the H(hsp/TP5) construct – one in the interior of the X chromosome – can enhance repression by the telomeric TP5 element. Enhancement of cytotype regulation can therefore be mediated by transgenic P elements at non-telomeric locations on any of the major chromosomes in the D. melanogaster genome.
(iii) Synergism between TPs and a transgene containing a mutant P coding sequence
Among the transgenes that interact with the telomeric elements TP5 and TP6, H(hsp/CP)2 has all three P introns, H(hsp/TP6)C has the first and the last intron, and H(hsp/TP5)D has the last intron. These three transgenes also contain the natural P element promoter. To determine if a transgene without any of the P introns or the P promoter could interact synergistically with the TPs, we tested TP5 and TP6 for their ability to repress GD in combination with H(hsp/P*)B, a transgene that contains a P-element coding sequence situated downstream of the hsp70 promoter. This transgene lacks the P promoter and all the P element's introns, and because of a frameshift mutation early in the P coding sequence, it cannot produce the functional P transposase. However, data from an RT–PCR experiment indicate that the P* element within H(hsp/P*)B is transcribed, even at 21 °C (Fig. 2). The hsp70 promoter in H(hsp/P*)B must therefore be functional even without a heat shock. The H(hsp/P*)B transgene is inserted on chromosome 3 close to the Rough eye (R) locus. Recombination data place it in the euchromatin between R and the telomere.

Fig. 2. RT–PCR analysis of H(hsp/P*) expression. RNA samples were extracted from groups of 20 females treated to three different conditions: 21 °C (held at this temperature until RNA extraction), 29 °C (held at 21 °C and then at this temperature overnight until RNA extraction) and HS (held at 21 °C and then subjected to a 45 min heat shock at 37 °C immediately before RNA extraction). Samples designated with a plus sign were reversed transcribed; those designated with a minus sign were not. The 1·8 kb product was generated by amplification with primers P▵0/1-d and P2575-u and the 1·4 kb product was generated by amplification with primers P▵0/1-d and P▵2/3-u.
The experiment to test for synergism between the telomeric elements and H(hsp/P*)B was initiated with reciprocal crosses between TP y+ w and w; H(hsp/P*)B strains. The TP y+ w/w; H(hsp/P*)B/ + F1 females were then crossed to Harwich y w males and their white-eyed and coloured-eyed F2 daughters were scored for GD; note that only two phenotypes segregated in these test crosses. To assess the repression abilities of H(hsp/P*)B and the TPs separately, we tested y w/y+ w; H(hsp/P*)B/ + F1 females from reciprocal crosses between w; H(hsp/P*)B and y w flies, and TP y+ w/y w F1 females from reciprocal crosses between TP y+ w and y w flies. The results of all these tests are summarized in Table 4 using GD frequencies that have been pooled over the segregating phenotypes.
Table 4. Synergism between the telomeric elements TP5 and TP6 and the H(hsp/P*)B transgene assessed in the F2 daughters of TP y+ w/w; H(hsp/P*)/+F1 females from reciprocal crosses between TP y+ w and w; H(hsp/P*)B strains
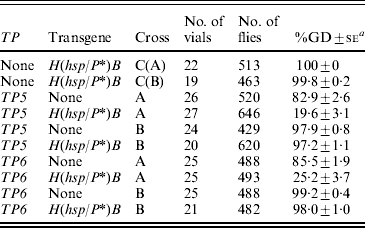
The initial crosses in this experiment were TP y +w females×y w males or y +w; H(hsp/P*)B males (cross A) and y w females or y +w; H(hsp/P*)B females×TP y +w males (cross B). The F1 females from these crosses were mated to Harwich y w males and their daughters were scored for GD. In the control crosses [C(A) and C(B)], the y w M strain was substituted for the TP strain. Females with and without the H(hsp/P*)B transgene were scored separately; however, because there were no significant differences between these two groups, the data have been pooled.
a Unweighted average percentage GD±se.
The crosses with H(hsp/P*)B alone (labelled as C(A) and C(B) in Table 4) showed that this transgene had no ability to repress GD. However, by themselves, both of the TPs were moderate repressors in cross A (83% GD with TP5 and 85% GD with TP6), but not in cross B (98 and 99% GD, respectively). In combination with the H(hsp/P*)B transgene, repression by the TPs was enhanced significantly in cross A (20 and 25% GD), but not in cross B (97 and 98% GD). Thus, in cross A, the H(hsp/P*)B transgene interacts synergistically with both the TPs to enhance cytotype regulation of the P-element family.
(iv) Collapse of synergistic repression when either the telomeric or transgenic P element is removed from the genotype
Stocks that contain TPs and many other dispersed P elements are powerful repressors of GD. When the TPs are removed from these stocks, repression ability persists, although much diminished, for at least one generation (Simmons et al., Reference Simmons, Niemi, Ryzek, Lamour, Goodman, Kraszkiewicz and Wolff2007). This lower level of repression has been explained by proposing that some of the other P elements are able to generate piRNAs, although much less vigorously than the major piRNA locus in the X telomere (Belinco et al., Reference Belinco, DiPrima, Wolff, Thorp, Buschette and Simmons2009). We tested whether or not repression could persist when a telomeric TP5 element is removed from a synergistic TP5-H(hsp/P) combination by measuring the repression ability of y w; H(hsp/TP5)D/+ females derived from crosses between TP5 y+ w/y w; H(hsp/TP5)D/+ mothers and y w fathers. The results of these tests, which spanned two generations, are summarized in Table 5.
Table 5. Collapse of synergistic repression of GD in the granddaughters of TP5 y+ w/y w; H(hsp/TP5)D/+ females
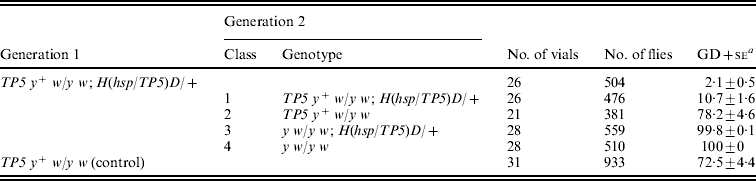
The females of generation 1 were produced by crossing TP5 y +w females with y w or y w; H(hsp/TP5)D males and the females of generation 2 were produced by crossing generation 1 females with y w males. Tests for repression of GD were conducted by crossing samples of generation 1 or generation 2 females to Harwich y w males. The data have been pooled over the genotypes that segregated in these crosses.
a Unweighted average percentage GD±se.
TP5 y+ w/y w; H(hsp/TP5)D/+ females in generation 1 produced daughters with a very low frequency of GD – only 2·1%; these females were, therefore, strong repressors of dysgenesis. By contrast, control TP5 y+ w/y w females of generation 1 produced daughters with a much higher GD frequency (72·5%). The difference between these two frequencies clearly demonstrates that the telomeric TP5 element and the H(hsp/TP5)D transgene interact synergistically to repress GD. This synergism persisted in the TP5 y+ w/y w; H(hsp/TP5)D/+ females of class 1 in generation 2 (10·7% GD), but was absent in their sisters that had lost either the H(hsp/TP5)D transgene (class 2, 78·2% GD), the telomeric TP5 element (class 3, 99·8% GD) or both (class 4, 100% GD). Removal of the H(hsp/TP5)D transgene therefore causes the repression mechanism – initially very strong – to collapse to what might be called its ‘ground state,’ and removal of the telomeric TP5 element causes it to collapse utterly.
(v) Impairment of synergistic repression by mutations in aub, piwi and Su(var)205
The proteins encoded by the genes aub and piwi play important roles in the piRNA pathway (Brennecke et al., Reference Brennecke, Aravin, Stark, Dus, Kellis, Sachidanandam and Hannon2007; Tushir et al., Reference Tushir, Zamore and Zhang2009), and the protein encoded by the Suppressor of variegation 205 [Su(var)205] gene – known as heterochromatin protein 1 (HP1) – plays an important role in chromatin organization (James et al., Reference James, Eissenberg, Craig, Diethrich, Hobson and Elgin1989). Mutational depletion of any of these proteins can impair P-element regulation profoundly. In particular, TPs in stocks that have been kept heterozygous for some aub, piwi or Su(var)205 mutations do not establish strong synergism with other P elements (Belinco et al., Reference Belinco, DiPrima, Wolff, Thorp, Buschette and Simmons2009). These same mutations would, therefore, be expected to prevent synergism between the telomeric TP5 element and the H(hsp/TP5)D transgene. To test this prediction, we crossed the H(hsp/TP5)D transgene into TP5 stocks that were heterozygous for aub, piwi or Su(var)205 mutations and evaluated the resulting TP5 y+ w/y w; mutation/H(hsp/TP5)D females for their ability to repress GD in the next generation. We also evaluated TP5 y+ w/y w; +/H(hsp/TP5)D females from parallel crosses in which the various mutations had been removed from the TP5 stocks many generations earlier.
As a negative control in this experiment, we crossed the H(hsp/TP5)D transgene into an M strain that did not have any P elements. When 24 females from this control were test crossed to Harwich w males, 98·6 ± 0·7% of their 345 daughters were dysgenic. Thus, by itself, the H(hsp/TP5)D transgene could not repress GD. As a positive control, we crossed the H(hsp/TP5)D transgene into a TP5 stock that was heterozygous for Gla, a mutation that has not been implicated in any aspect of piRNA-mediated regulation; this stock was the source of the telomeric TP5 element in all the other mutant stocks. When 25 females from this control were test crossed to Harwich w males, 17·0 ± 2·1% of their 486 daughters were dysgenic. Thus, when H(hsp/TP5)D was combined with TP5 from the root Gla stock, GD was repressed strongly.
The results of the tests with the other mutant and mutant-free TP5 stocks are summarized in Table 6. In general, when H(hsp/TP5)D was crossed into the TP5 stocks from which the aub, piwi and Su(var)205 mutations had been removed, dysgenesis was repressed strongly (9·6–19·4% GD), as in the positive control. The only exception was the stock from which piwi 2 had been removed, where the GD frequency was 52·4%. This higher frequency does not appear to be related to any long-term effect of the piwi 2 mutation; rather, it may simply be due to a random change in the structure and properties of the XL telomere (Belinco et al., Reference Belinco, DiPrima, Wolff, Thorp, Buschette and Simmons2009).
Table 6. Effects of aub, piwi and Su(var)205 mutations on synergism between the telomeric TP5 element and the H(hsp/TP5)D transgene
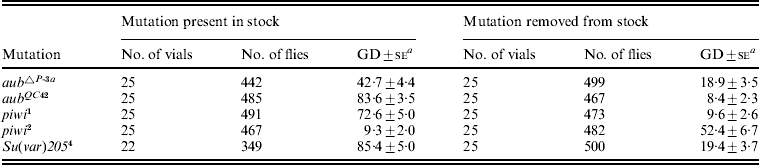
TP5 y +w females from stocks with and without the listed mutations (Belinco et al., Reference Belinco, DiPrima, Wolff, Thorp, Buschette and Simmons2009) were crossed to y w; H(hsp/TP5)D males to produce TP5 y +w/y w; (mutation)/H(hsp/TP5)D females, which were then tested for repression of GD by crosses to Harwich w males. Daughters with different TP5 and H(hsp/TP5)D genotypes were not scored separately.
a Unweighted average percentage GD±se.
When H(hsp/TP5)D was crossed into the TP5 stocks that were heterozygous for the various mutations, GD was generally not repressed strongly (42·7–85·4% GD). The only exception was the stock heterozygous for piwi 2, where the GD frequency was 9·3%. This lower frequency may reflect that piwi 2 is a weaker mutant allele than piwi 1; for instance, in homozygous condition piwi 2 causes female sterility, whereas homozygous piwi 1 also causes male sterility. A comparison of the left and right sides of Table 6 indicates that synergism between TP5 and H(hsp/TP5)D was impaired when the TP5 element came from a stock that was heterozygous for aub▵P − 3a, aubQC42, piwi 1 or Su(var)205 4. These results are similar to those from tests for synergism between TP5 and an ensemble of non-TPs (Belinco et al., Reference Belinco, DiPrima, Wolff, Thorp, Buschette and Simmons2009). The potential for synergism between TP5 and another P element – for example, the one in the H(hsp/TP5)D transgene – therefore appears to be sensitive to the mutational depletion of proteins encoded by the aub, piwi and Su(var)205 genes. It is important to recognize that this conclusion is based on tests with TP5 stocks that were heterozygous for the various mutations, and that in these stocks the capacity for synergism may have been impaired by the long-term effects of the mutations on the function of the TP5 element within the XL telomere. However, this impairment is not permanent because when TP5 stocks from which the mutations were removed many generations earlier are tested for synergism with H(hsp/TP5)D, dysgenesis is once again repressed strongly. The negative effects of the mutations can therefore be reversed after the mutations have been removed from the stocks.
4. Discussion
P elements provide an unusual opportunity to elucidate the mechanisms that regulate eukaryotic transposons. Individual P elements can be isolated in an otherwise P-element-free genotype and then assessed for their abilities to prevent hybrid dysgenesis in test crosses. The regulatory abilities of combinations of P elements can also be assessed. These experimental tests are specific for P-element activity in the germ line and yield quantitative data to document it. The experimental end-points – for example, the frequency of GD in the offspring of a test cross – therefore directly reveal whether or not particular P elements, or combinations of P elements, are able to prevent P excision and transposition in the germ line. No other transposon affords the possibility of defining and controlling the genotype so precisely, and of connecting it to quantitative data on transposition.
We have used genetic manipulations to determine if the regulatory abilities of TPs – which are anchors of the P cytotype – can be enhanced by other P elements at non-telomeric locations. The telomeric elements TP5 and TP6, both inserted in the TAS of chromosome XL, repress GD in their own right, presumably because they are situated in a major locus for the production of piRNAs. Other P elements in the XL TAS have been shown to repress GD (Ronsseray et al., Reference Ronsseray, Lehmann and Anxolabéhère1991; Marin et al., Reference Marin, Lehmann, Nouaud, Izaabel, Anxolabéhère and Ronsseray2000), and their piRNA output has been documented (Brennecke et al., Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008). Furthermore, these piRNAs are deposited maternally in eggs (Brennecke et al., Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008), which is consistent with the abilities of X-linked TPs to repress GD through strictly maternal effects. Maternally deposited small RNAs have also been implicated in the repression of dysgenesis induced by the Penelope transposon in D. virilis (Blumenstiel & Hartl, Reference Blumenstiel and Hartl2005).
We have shown that the regulatory abilities of TP5 and TP6 are markedly enhanced by different types of transgenic P elements inserted at non-telomeric locations on the X chromosome or on either of the major autosomes. In their own right, none of these transgenic P elements has any ability to repress GD. The enhancement of regulatory ability must therefore be due to synergism between the telomeric and transgenic P elements, not to the addition of separate regulatory effects. Previous work had shown that TP5 and TP6 repress dysgenesis synergistically when combined with an ensemble of heterogeneous, dispersed, non-TPs (Simmons et al., Reference Simmons, Niemi, Ryzek, Lamour, Goodman, Kraszkiewicz and Wolff2007; Belinco et al., 2009). We now know that the regulatory abilities of these TPs can be enhanced synergistically by a single transgenic P element.
Both small and large transgenic P elements enhanced the regulatory abilities of the TPs. The small enhancing elements were transgenic clones of TP5 and TP6. Each of these transgenic TPs was effective in boosting the regulatory ability of each of the native TPs. However, with both of the native TPs, the transgenic TP5 element was a more effective enhancer than the transgenic TP6 element. That the transgenic TP5 should be more effective when combined with its cognate telomeric TP5 is perhaps not surprising because these two elements are perfectly identical (except for the terminal truncations in the transgenic construct). However, the transgenic TP5 element was also a better enhancer in combination with the telomeric TP6 element, with which it shares only 83% of its sequence. Thus, regulatory enhancement is not simply a function of the amount of sequence shared by the telomeric and transgenic P elements. Other features of the elements, such as their expression level, their specific sequence composition, or the ease with which their RNA products are transported within and between cells, could be relevant. However, some minimum amount of shared sequence appears to be needed for synergism because the very small transgenic SP element did not enhance regulation when it was combined with either of the TPs.
The large transgenic P elements CP and P* both boosted regulation with each of the TPs. CP encodes the P transposase and might be expected to exacerbate dysgenesis. However, when combined with either TP, it led to substantially less dysgenesis in the test cross offspring. P* is a frame-shifted P coding sequence minus the native P promoter and all three P introns. When positioned downstream of the hsp70 promoter in a hobo transgene, this element also enhanced TP-anchored regulation significantly. Thus, regulatory synergism occurs even when the transgenic P element encodes the transposase or when it lacks the native P promoter and all three P introns. It is interesting, however, that neither the CP nor P* transgenic elements was as effective as the transgenic TP5 element in boosting regulation. Both of the TPs share most of their sequence with these two transgenic P elements. Thus, as discussed above, shared sequence is not the sole determiner of enhanced regulation.
Previous studies have shown that a TP's ability to repress GD is transmitted to test cross offspring as a strictly maternal effect – that is, offspring that do not inherit the TP repress GD as well as those that do inherit it (Thorp et al., Reference Thorp, Chapman and Simmons2009; Belinco et al., 2009; Simmons et al., Reference Simmons, Raymond, Niemi, Stuart and Merriman2010). This observation indicates (1) that repression involves a product of the TP, not the TP itself, (2) that the amount of product transmitted through the egg is sufficient to repress GD in the zygote, although perhaps not in every zygote and (3) that if any more TP product is synthesized in the zygote, it does not make repression any stronger. Thus, the final level of repression appears to be established – that is, set – in the maternal germ line. These conclusions also hold when repression is enhanced by combining a transgenic P element with a TP in the mother's genotype. Test cross offspring that inherit neither transgenic P nor TP repress GD as well as offspring that inherit both. Cases in which the maternal genotype brings about strong, but incomplete, repression are particularly interesting. For example, when H(hsp/CP)2 is combined with either of the TPs the GD frequency in the test cross offspring is around 20%, regardless of the offspring's genotype (Tables 1 and 2). Offspring that inherit the TP, H(hsp/CP)2, or both are not better at repressing GD than offspring that inherit neither of these factors even though there is clearly ‘room for improvement’. The enhanced regulatory state, like the basal regulatory state, therefore appears to be set in the maternal germ line.
One other feature of the experimental data is that the level of repression in the test cross offspring is strongly influenced by the grand-parental origin of the TP. When the TP is derived from the grandmother and the transgenic P element from the grandfather, repression is much stronger than when the derivation is reversed. TPs that are paternally derived completely lose their regulatory power (Stuart et al., Reference Stuart, Haley, Swedzinski, Lockner, Kocian and Simmons2002; Simmons et al., Reference Simmons, Raymond, Niemi, Stuart and Merriman2004). However, these elements can be ‘resuscitated’ if they pass through the germ line of a daughter (Niemi et al., Reference Niemi, Raymond, Patrek and Simmons2004). From the data in Tables 1 and 2, it appears that resuscitation is facilitated by the presence of a transgenic P element in the daughter's genotype.
What molecular mechanisms underlie these phenomena? Cytotype regulation appears to be mediated by piRNAs generated from P elements inserted in the TAS of chromosome XL (Brennecke et al., Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008). The biogenesis of these RNAs from the TPs is not understood. However, once formed, it is thought that the piRNA population is amplified by a ping-pong cycle fed by antisense RNAs transcribed from the TP and sense RNAs transcribed from other P elements (Brennecke et al., Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008; Belinco et al., 2009). Our data are consistent with this hypothesis. P elements contained within hobo transgenes clearly strengthen the regulatory abilities of TPs, presumably by providing the sense transcripts needed to amplify P-specific piRNAs in the maternal germ line. Ping-pong amplification of piRNAs is thought to occur in the nuage, a region on the cytoplasmic side of the nuclear membrane in germ line cells (Lim & Kai, Reference Lim and Kai2007; Kibanov et al., Reference Kibanov, Egorova, Ryanzansky, Sokolova, Kotov, Olenkina, Stolyarenko, Gvozdev and Olenina2011; Nagao et al., Reference Nagao, Sato, Nishida, Siomi and Siomi2011; Zhang et al., Reference Zhang, Xu, Koppetsch, Wang, Tipping, Ma, Weng, Theurkauf and Zamore2011; Anand & Kai, Reference Anand and Kai2012). Several proteins implicated in ping-pong cycling have been localized to the nuage. It is possible that P-element transcripts exported from germ-line nuclei could be processed into piRNAs by these proteins, particularly in the nurse cells, which could, in turn, export them to the developing oocyte where they would accumulate to provide a defence against P activity in the future embryo. Synergism between the telomeric and transgenic P elements is therefore consistent with an important role for ping-pong cycling in cytotype regulation.
Synergism might also be explained by transcription of P mRNAs by an RNA-dependent RNA polymerase (RdRP), generating antisense P RNAs that might feed into a pathway for the production of small interfering RNAs (siRNAs). However, we found that cytotype enhancement is impaired by mutations in the aub and piwi genes, both of which encode proteins that bind piRNAs. In addition, a telomeric P trans-silencing effect is impaired by mutations in these two genes, as well as by mutations in other genes known to be involved in the piRNA pathway; however, it is not impaired by mutations in r2d2, a gene in the siRNA pathway, or in loquacious, a gene in the miRNA and endo-siRNA pathways (Josse et al., Reference Josse, Teysset, Toideschini, Sidor, Anxolabéhère and Ronsseray2007; Todeschini et al., Reference Todeschini, Teysset, Delmarre and Ronsseray2010). These findings argue that cytotype regulation is mediated by piRNAs rather than siRNAs. Furthermore, there is currently no evidence for an RdRP in Drosophila. The amplification of piRNAs by ping-pong cycling therefore appears to be the more plausible explanation for how non-TPs enhance TP-anchored cytotype regulation in the D. melanogaster germ line.
Jordan Becker prepared Fig. 1 and Donald Rio provided a clone containing the frameshifted P-element coding sequence P*. Johng Lim made helpful comments on an early version of the manuscript and Justin Blumenstiel read the final manuscript and suggested some improvements. The experimentation was supported by funds from the Department of Genetics, Cell Biology, and Development of the University of Minnesota. Construction of the H(hsp/P*) transgene was accomplished under the auspices of a past grant from the National Institutes of Health.










