Introduction
Psychiatric disorders place an enormous burden on medical resources and society in general (Eaton et al., Reference Eaton, Martins, Nestadt, Bienvenu, Clarke and Alexandre2008; Petrou et al., Reference Petrou, Johnson, Wolke, Hollis, Kochhar and Marlow2010). For most disorders, the causal factors are as yet largely unknown which limits treatment options considerably. A better understanding of the etiology of psychiatric disorders is a crucial step towards advancing treatment and intervention strategies.
Twin studies showed that genetic factors play an important role in the etiology of psychiatric traits. Heritability estimates (h2, i.e. the inherited contribution of genetic variance to trait variance) range from 35% for major depression to over 60% for schizophrenia (SCZ) (Polderman et al., Reference Polderman, Benyamin, de Leeuw, Sullivan, van Bochoven, Visscher and Posthuma2015). The remaining variance is explained by non-genetic factors perhaps including non-identifiable environmental factors. Another method to derive estimates of genetic and environmental variance is the use of pedigree data (e.g. parents and children, siblings and half-siblings) from large national registers (Pettersson et al., Reference Pettersson, Larsson and Lichtenstein2016). Additionally, rapid methodological developments have recently advanced the use of summary data of genome-wide association studies (GWAS) in which heritability is inferred from the linkage disequilibrium scores (LDSC) of single nucleotide polymorphisms (SNP) (i.e. the ability of a SNP to tag other genetic variants) (Bulik-Sullivan et al., Reference Bulik-Sullivan, Loh, Finucane, Ripke, Yang, Patterson, Daly, Price and Neale2015). Yet, it is recognized that these heritability estimates do not capture all genetic factors contributing to variance in the trait (such as rare genetic effects), and hence can be viewed as lower bound estimates.
While heritability is conceptualized as a single population parameter, estimates depend on the population under study, ascertainment, diagnostic instruments, and study design. Estimates may also change over time due to variations in diagnostic criteria (Zablotsky et al., Reference Zablotsky, Black, Maenner, Schieve and Blumberg2015; Thomas et al., Reference Thomas, Sanders, Doust, Beller and Glasziou2015), an increase in awareness and detection of psychiatric disorders (Van Naarden Braun et al., Reference Van Naarden Braun, Christensen, Doernberg, Schieve, Rice, Wiggins, Schendel and Yeargin-Allsopp2015), changes in the exposure to environmental factors (Rokholm et al., Reference Rokholm, Silventoinen, Tynelius, Gamborg, Sørensen and Rasmussen2011), or changes in social situations (Kendler et al., Reference Kendler, Thornton and Pedersen2000).
Using two different methods, this study capitalizes on the largest and most powerful data-sets to date, to estimate the heritability of eight psychiatric conditions: (1) alcohol dependence (AD), (2) anorexia nervosa (AN), (3) attention deficit/hyperactivity disorder (ADHD), (4) autism spectrum disorder (ASD), (5) bipolar disorder (BIP), (6) major depressive disorder (MDD), (7) obsessive-compulsive disorder (OCD), and (8) SCZ. First, we use a large Swedish national cohort (h2-national) that currently includes over 20 million full and maternal half-sibling pairs. Unlike most twin studies, the Swedish sibling sample uses clinical diagnoses derived from medical in- and out-patient treatment registers, instead of surveys. Second, we use summary data of eight large samples of subjects with measured SNPs (h2-SNP). The uniqueness is estimating heritability from very large samples, based on genetic similarities inferred from distantly related people. As in the h2-national design, case status in the h2-SNP design is based on diagnostic criteria.
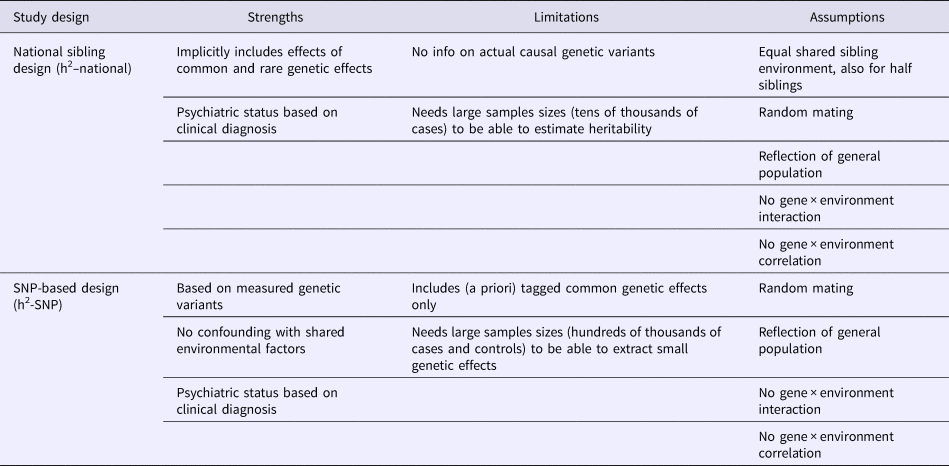
Although each study design has its own strengths, they also have study-specific biases and assumptions (listed in Table 1). For instance, in the national register data that we use, not all affected individuals seek help or are correctly diagnosed. The sibling method also relies on certain assumptions, such as a 100% shared environment despite age differences between siblings, and despite the fact that half-siblings might live in two families (with the biological mother and with the biological father) and thus spend potentially less time together than full siblings (Moffitt et al., Reference Moffitt, Caspi, Taylor, Kokaua, Milne, Polanczyk and Poulton2010; Pettersson et al., Reference Pettersson, Larsson and Lichtenstein2016). However, confounding with shared environmental factors is likely excluded in the h2-SNP design. Yet, in contrast to sibling studies that assume to capture all possible genetic effects, the h2-SNP analysis is based on genome-wide variation that is derived from a selection of common genetic variants only (Vinkhuyzen et al., Reference Vinkhuyzen, Wray, Yang, Goddard and Visscher2013). By including a large number of observations from both study designs, our study is adequately powered to robustly estimate in each design the heritability, despite relying on different sets of assumptions and methodologies.
Table 1. Strengths, limitations and assumptions of each study design in estimating heritability (h2) of psychiatric disorders
The aim of this study is to provide a test of the homogeneity in heritability estimates between family-based data (h2-national) and SNP-based data for eight psychiatric conditions. Our hypothesis is that h2-SNP is lower, but correlates positively with the family-based estimates. In addition, our design can illustrate the differences in etiology between the psychiatric conditions, guiding future directions for etiological research in psychiatry.
Method
National sibling cohort (h2-national)
Personal identification numbers unique to each individual in Sweden were used to create a national population-based cohort from which twins were excluded. Information was extracted from the National Patient Register, which includes all public psychiatric inpatient diagnoses in Sweden since 1973 and outpatient diagnoses since 2001, assigned by the attending physician with a non-hierarchical diagnostic structure in accord with ICD version 8 (1969–1986), 9 (1987–1996), or 10 (1997–present). The ICD codes for the eight disorders are presented in Table S1 in the Supplementary. We used the Multi-Generation Register to link individuals to their full and maternal half-siblings registered as living in Sweden since 1961 and born in Sweden since 1932. Only two siblings per family were included, starting with the oldest siblings in each family followed by the next oldest sibling, but only if born within 5 years of the first sibling to maximize the probability that they had experienced a similar rearing (i.e. shared) environment. If the two eldest were born more than 5 years apart, we proceeded to the second oldest sibling pair within the family, and so on. The final sample size of full- and maternal half-siblings varied by diagnosis. To ensure that the younger sibling in each pair had lived long enough to receive a potential diagnosis, pairs in which the younger sibling was younger than 5 (for ADHD and ASD), 10 (for AD, AN, MDD, and OCD), or 15 years old (BIP and SCZ) were excluded.
The sibling design assumes (a) that full siblings share an average of 50% of additive genetic effects and 25% of non-additive genetic effects, (b) that maternal half-siblings share an average of 25% of additive genetic effects and 0% of non-additive genetic effects, and (c) that shared environmental effects are 100% shared between both full and maternal half-siblings, and that (d) non-shared environmental effects are unique to each individual. By comparing the observed tetrachoric correlations between the binary diagnoses for full and maternal half-siblings, we estimated the contribution of genetic variance (h2-national), and shared and non-shared environmental variance. The analyses were carried out in Mplus (Muthén and Muthén, Reference Muthén and Muthén1998) using the mean- and variance-adjusted unweighted least squares estimator. We regressed out the effects of sex and age from all diagnoses. For ADHD and ASD, we limited the birth year to 1990 and beyond because these diagnoses only existed in ICD 9 and 10.
Genetic data (h2-SNP)
The h2-SNP estimates were based on the most recent available data for all eight disorders in the Psychiatric Genomics Consortium (PGC)(Psychiatric GWAS Consortium Coordinating Committee et al., Reference Cichon, Craddock, Daly, Faraone, Gejman, Kelsoe, Lehner, Levinson, Moran, Sklar and Sullivan2009; Psychiatric GWAS Consortium Steering Committee, 2009) (see Study cohort details in the Supplementary). The LDSC approach was used to estimate the h2-SNP (Bulik-Sullivan et al., Reference Bulik-Sullivan, Loh, Finucane, Ripke, Yang, Patterson, Daly, Price and Neale2015). In brief, this method is based on the LDSC of a SNP, which reflects its ability to tag other SNPs. The more SNPs are tagged, the higher the probability that this represents a polygenic signal instead. Therefore, by taking LD into account this method is able to distinguish spurious associations due to population stratification from the true polygenic signal. The LDSC method is also robust to confounding due to shared environmental effects, and is very efficient as it can be applied to GWAS summary statistics (Evans et al., Reference Evans, Tahmasbi, Vrieze, Abecasis, Das, Gazal, Bjelland, de Candia, Goddard, Neale, Yang, Visscher and Keller2018). Of note, GWAS usually include millions of common variants, but not rare variants. The heritability (h2-SNP), adjusted for the prevalence of the disorder, is inferred from the slope of the regression.
Analyses
Differences in heritability estimates were tested using d = h12 − h22, ![]() $\hbox{S}{\rm. E}{\rm.} d = \sqrt {{(\hbox{S}{\rm. E}{\rm.} \hbox{h}_1^2 )}^2 + {(\hbox{S}{\rm. E}{\rm.} \hbox{h}_2^2 )}^2} $. The ratio Z = d/s.e.(d) gives a test of the null hypothesis that the difference d is zero, by comparing the value of Z to the standard normal distribution(Altman and Bland, Reference Altman and Bland2003). A significance threshold of 0.05 was Bonferroni corrected to accommodate multiple testing (i.e. a correction for eight tests).
$\hbox{S}{\rm. E}{\rm.} d = \sqrt {{(\hbox{S}{\rm. E}{\rm.} \hbox{h}_1^2 )}^2 + {(\hbox{S}{\rm. E}{\rm.} \hbox{h}_2^2 )}^2} $. The ratio Z = d/s.e.(d) gives a test of the null hypothesis that the difference d is zero, by comparing the value of Z to the standard normal distribution(Altman and Bland, Reference Altman and Bland2003). A significance threshold of 0.05 was Bonferroni corrected to accommodate multiple testing (i.e. a correction for eight tests).
Results
Estimates of shared environmental effects were non-significant in the sibling analyses. The heritability estimates (Fig. 1) showed significant differences between the h2-national estimates and the h2-SNP estimates. The latter were significantly lower (corrected p < 0.02), except for AN, BIP, and OCD where the h2-SNP estimates did not significantly differ from h2-national (corrected p > 0.30). However, these differences should be interpreted with caution as due to the somewhat smaller samples sizes of these particular disorders, the standard errors (s.e.) of h2-national were relatively wide for AN, BIP, and OCD. Of note, the s.e. is in general sensitive to sample size, and in particular for h2-national because the full and half-sibling groups only differ by 0.25 in genetic relatedness. Additionally, the nature of summary data of large consortium designs implies that included samples have been genotyped on different platforms and chips, potentially increasing the s.e. of h2-SNP.
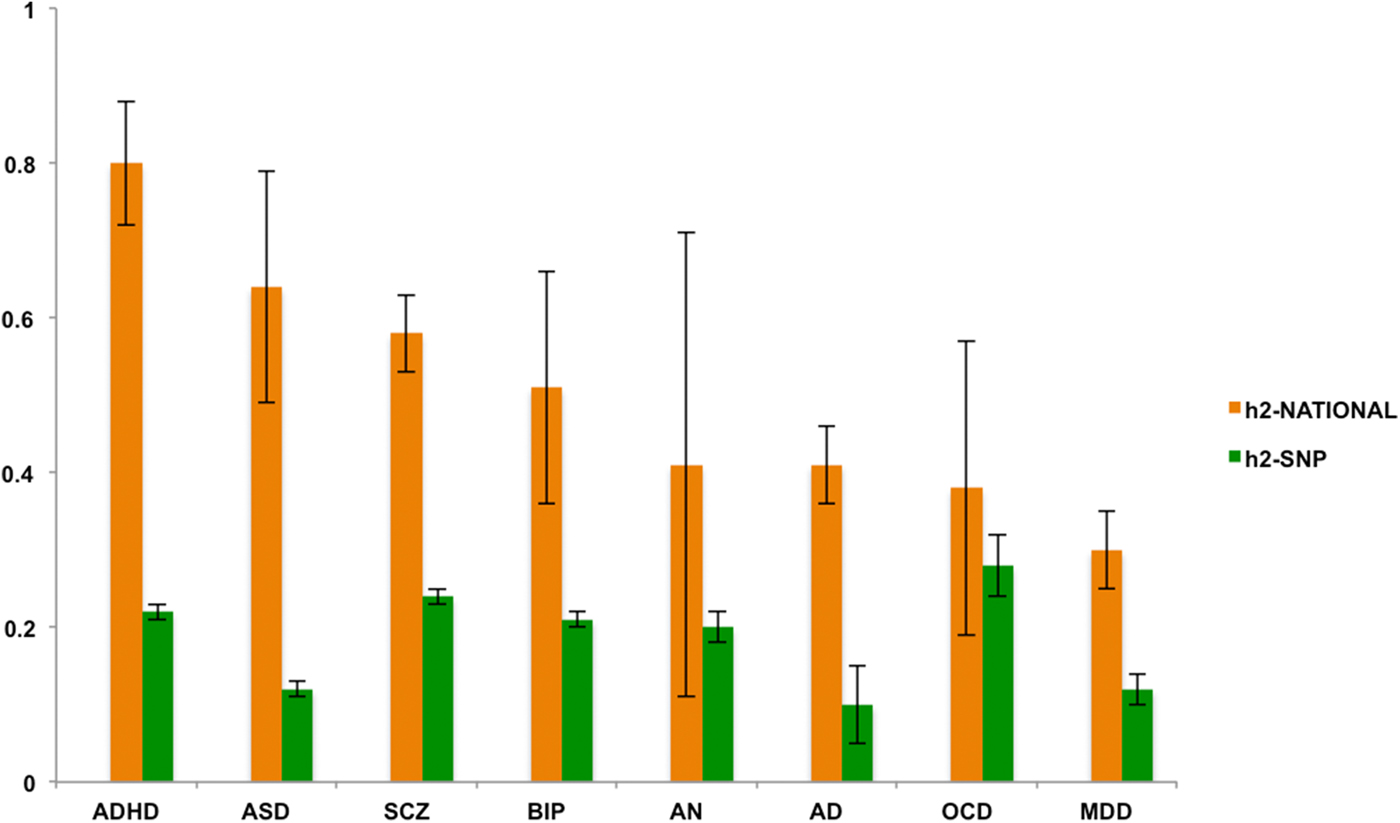
Fig. 1. h2-national and h2-SNP estimates ordered from low to high based on h2-national. Note: Error bars represent standard errors. AD, alcohol dependence; ADHD, attention deficit/hyperactivity disorder; AN, anorexia nervosa; ASD, autism spectrum disorder; BIP, bipolar disorder; MDD, major depressive disorder; OCD, obsessive-compulsive disorder; SCZ, schizophrenia.
Heritability estimates from the two designs correlated positively (r 0.19). However, this correlation was mainly driven by OCD that showed the highest h2-SNP and lowest h2-national; when removing OCD this correlation increased to 0.50. The high h2-SNP is probably due to the fact that the OCD sample is heavily ascertained from highly multiplex families and early age of onset cases, and consists thus of the most severe and genetically loaded cases.
Tables S2 and S3 in the Supplementary show detailed sample characteristics of each study design.
Discussion
We estimated the heritability of eight psychiatric conditions using two different study designs: a national sibling design, and a SNP-based design. Estimates derived from the two study designs consistently show that all disorders are moderate to highly heritable but also showed large differences between disorders. The correlation between the family-based and SNP-based estimates was positive (0.50 when leaving out OCD) suggesting that a higher family-based heritability is associated with a larger (aggregated) effect of SNPs. This supports the hypothesis that, apart from rare variants, common genetic variants play an important role in psychiatric disorders, and thus confirms the polygenic nature of these complex traits (Visscher et al., Reference Visscher, Goddard, Derks and Wray2012).
The heritability estimates based on the large national sibling study (h2-national) were remarkably similar to previous twin studies of psychiatric traits (Polderman et al., Reference Polderman, Benyamin, de Leeuw, Sullivan, van Bochoven, Visscher and Posthuma2015), despite different assessment strategies, with twin studies being survey-based, and as such based on psychiatric trait measures, and the national sibling study based on clinical diagnoses. This might suggest that heritability estimates are robust across different diagnostic tools and measures. It is also in line with studies that reported high genetic correlations between survey-based psychiatric traits and clinical diagnoses, e.g. for ASD (Colvert et al., Reference Colvert, Tick, McEwen, Stewart, Curran, Woodhouse, Gillan, Hallett, Lietz, Garnett, Ronald, Plomin, Rijsdijk, Happé and Bolton2015), ADHD (Lubke et al., Reference Lubke, Hudziak, Derks, van Bijsterveldt and Boomsma2009), and psychosis (Zavos et al., Reference Zavos, Freeman, Haworth, McGuire, Plomin, Cardno and Ronald2014), suggesting an overlap in genetic factors between psychiatric traits as measured in the general population and clinical disorders.
Heritability estimates based on SNP data (h2-SNP) were, as expected, lower than the family-based designs (Yang et al., Reference Yang, Zeng, Goddard, Wray and Visscher2017). An obvious explanation for these differences is that h2-SNP is based on measured common (and not rare) genotypes, whereas the other design is based on familial relationships and hence includes estimates of genetic factors shared by relatives that are rare in populations. The largest differences between the family-based and h2-SNP estimates were observed for the neurodevelopmental traits ADHD and ASD, and for SCZ. Indeed, rare variant risk effects have been reported for ADHD, ASD, and SCZ (Williams et al., Reference Williams, Zaharieva, Martin, Langley, Mantripragada, Fossdal, Stefansson, Stefansson, Magnusson, Gudmundsson, Gustafsson, Holmans, Owen, O'Donovan and Thapar2010; Hiroi et al., Reference Hiroi, Takahashi, Hishimoto, Izumi, Boku and Hiramoto2013; Sanders et al., Reference Sanders, He, Willsey, Ercan-Sencicek, Samocha, Cicek, Murtha, Bal, Bishop, Dong, Goldberg, Jinlu, Keaney, Klei, Mandell, Moreno-De-Luca, Poultney, Robinson, Smith, Solli-Nowlan, Su, Teran, Walker, Werling, Beaudet, Cantor, Fombonne, Geschwind, Grice, Lord, Lowe, Mane, Martin, Morrow, Talkowski, Sutcliffe, Walsh, Yu, Ledbetter, Martin, Cook, Buxbaum, Daly, Devlin, Roeder and State2015), although a recent well-powered study on SCZ showed that the explained variance due to rare variants was about 20% of the total explained variance (0.85% for rare variants v. 3.4% for common variants) (CNV and Schizophrenia Working Groups of the Psychiatric Genomics Consortium & Psychosis Endophenotypes International Consortium, 2017).
Another explanation for the discrepancy between family-based and SNP-based estimates for ADHD, ASD, and SCZ could be the presence of non-additive effects resulting in overestimates of the narrow-sense heritability in the family-based design, when non-additive influences are removed from the statistical models. Non-additive factors contributing to trait variance have indeed been reported for ADHD (Rietveld et al., Reference Rietveld, Hudziak, Bartels, van Beijsterveldt and Boomsma2004). Lower h2-SNP might also indicate the presence of disorder heterogeneity, that is, a disorder is viewed as a single disorder but actually being a combination of disorder dimensions that has biologically distinct causal factors. As this affects GWAS, and hence h2-SNP, most substantially, this explains the lower h2-SNP but is also informative about potentially underlying disorder mechanisms (Wray and Maier, Reference Wray and Maier2014). For ADHD and ASD specifically, the inclusion of trio data (i.e. case-pseudo control design) may have underestimated h2-SNP due to an increased polygenic burden on the un-transmitted chromosomes (Peyrot et al., Reference Peyrot, Boomsma, Penninx and Wray2016a), although the trio samples were small compared with the much larger case-control samples.
In general, the nature of the large consortium designs on which SNP-based heritability is based likely increases the standard error, all of which will impact on h2-SNP. The h2-SNP of the eight psychiatric traits as observed in the current study should, therefore, be considered as lower-bound estimates of SNP-heritability. Interestingly, the smallest difference in family-based and SNP-based estimates between both designs was for OCD (respectively, 0.38% v. 0.28%). As mentioned previously, the SNP-based estimate of OCD was based on a clinical sample of most severe and therefore probably most genetically loaded cases. Yet, standard errors for both the family-based and SNP-based estimate of OCD were relatively large, so these findings should be interpreted with caution. In a similar vein, one should not stretch the interpretation of the SNP based AD estimate as it derives from one of the smaller genetic samples.
The family-based estimates showed substantial differences in the relative contributions of genes and environment across the eight psychiatric conditions: Heritability estimates for AD, AN, MDD, and OCD were relatively low, ranging from 30 to 41%. However, the prevalence of AN in the family data was low and hence, statistical power was limited, as illustrated by the large standard error in these data. Still, the estimate of 41% for AN in the family data confirms heritability estimates based on twin studies (Polderman et al., Reference Polderman, Benyamin, de Leeuw, Sullivan, van Bochoven, Visscher and Posthuma2015), also in clinical samples (Mazzeo et al., Reference Mazzeo, Mitchell, Bulik, Reichborn-Kjennerud, Kendler and Neale2009). Heritability estimates of ADHD, ASD, BIP, and SCZ showed the highest narrow-sense heritability estimates between 51 and 80%. With the dramatic increase in sample sizes, the recent endeavors to identify genes that could explain the heritability of psychiatric disorders is becoming more successful. For instance, 108 significantly associated genetic loci were identified for SCZ in a sample of almost 37 000 cases and over 113 000 controls (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). Follow-up analyses on biological pathways revealed that some of the associated genes play an important role in the immune system. However, high heritability not necessarily implies that genetic associations are easy to detect. For example, a recent study in 16 539 ASD cases and over 150 000 controls resulted in only one associated genetic locus (Warrier et al., Reference Warrier, Grasby, Uzefovsky, Toro, Smith, Chakrabarti, Khadake, Mawbey-Adamson, Litterman, Hottenga, Lubke, Boomsma, Martin, Hatemi, Medland, Hinds, Bourgeron and Baron-Cohen2017). Similarly, for ADHD only very recently the first 12 associated genetic loci have been published (Demontis et al., Reference Demontis, Walters, Martin, Mattheisen, Als, Agerbo, Belliveau, Bybjerg-Grauholm, Bækved-Hansen, Cerrato, Chambert, Churchhouse, Dumont, Eriksson, Gandal, Goldstein, Grove, Hansen, Hauberg, Hollegaard, Howrigan, Huang, Maller, Martin, Moran, Pallesen, Palmer, Pedersen, Pedersen, Poterba, Poulsen, Ripke, Robinson, Satterstrom, Stevens, Turley, Won, Andreassen, Burton, Boomsma, Cormand, Dalsgaard, Franke, Gelernter, Geschwind, Hakonarson, Haavik, Kranzler, Kuntsi, Langley, Lesch, Middeldorp, Reif, Rohde, Roussos, Schachar, Sklar, Sonuga-Barke, Sullivan, Thapar, Tung, Waldman, Nordentoft, Hougaard, Werge, Mors, Mortensen, Daly, Faraone, Børglum and Neale2017), illustrating that gene identification in the psychiatric domain is a long and complex avenue.
The fact that heritability estimates show variation across disorders and is never estimated >90% indicates that, discounting potential measurement error, stochasticity, and non-definable environmental factors, definable environmental factors might play an important role in the etiology of psychiatric disorders. In particular, for AD, AN, MDD, and OCD with heritability estimates <50%, environmental etiological research might elucidate crucial pathways that significantly increase the risk for these traits. In addition, gene by environment interaction will likely play a role in the development of psychiatric disorders (Uher and Zwicker, Reference Uher and Zwicker2017). For instance, one study showed that the effect of a genetic risk for MDD increased in individuals with childhood trauma (Peyrot et al., Reference Peyrot, Milaneschi, Abdellaoui, Sullivan, Hottenga, Boomsma and Penninx2014). In other words, given a genetic vulnerability, exposure to certain environmental risk factors will increase the risk for disorder development. However, two recent larger studies (Mullins et al., Reference Mullins, Power, Fisher, Hanscombe, Euesden, Iniesta, Levinson, Weissman, Potash, Shi, Uher, Cohen-Woods, Rivera, Jones, Jones, Craddock, Owen, Korszun, Craig, Farmer, McGuffin, Breen and Lewis2016; Peyrot et al., Reference Peyrot, Van der Auwera, Milaneschi, Dolan, Madden, Sullivan, Strohmaier, Ripke, Rietschel, Nivard, Mullins, Montgomery, Henders, Heat, Fisher, Dunn, Byrne, Air, Baune, Breen, Levinson, Lewis, Martin, Nelson, Boomsma, Grabe, Wray and Penninx2017) showed no interaction effect. All in all, the empirical evidence for gene by environment interaction in psychiatric disorders is as yet limited (Wray and Maier, Reference Wray and Maier2014) but increasing sample sizes and careful assessments of environmental risk factors are crucial in future research aiming to elucidate causal routes in psychiatric disorders.
Limitations
Our study has potential limitations. First, the concept of shared environment is not straightforward in the sibling design. Siblings are born at different points in time, do not share the prenatal environment, and are born into different family structures (e.g. the first child born to young parents v. the second child born to older parents who already have one child). We aimed to minimize potential time effects by limiting age differences between siblings to a maximum of 5 years. Moreover, additional analyses comparing siblings born within 1–2 v. 4–5 years apart showed very similar results (data not shown). Second, in the national sibling design, we assume that the shared environment of full and half siblings is the same. This assumption seems correct: A recent study showed that the vast majority of both Swedish full and maternal half-siblings tend to live together throughout childhood (Pettersson et al., Reference Pettersson, Larsson and Lichtenstein2016). Third, the inclusion age limit for the different disorders in the national sibling design was relatively young (e.g. minimum age of 10 years old for AD, AN, MDD, and OCD) to strike a balance between power on the one hand, and clinical generalizability on the other. However, we cannot rule out that children of that age develop such a disorder later in life. Yet, two additional sets of sensitivity analyses in which only older subjects were included, showed very similar results (online Tables A1 and A2). Fourth, the national sibling cohort lacked information from primary care, which might result in false negatives, in particular, for disorders from the internalizing spectrum and drug abuse (Sundquist et al., Reference Sundquist, Ohlsson, Sundquist and Kendler2017). However, this source of bias probably has limited influence on the heritability estimates as it is unlikely to differentially impact full- v. maternal half-siblings. Nevertheless, failure to include information from primary care decreases power, and limits the generalizability of the study results to the more severe forms of mental health problems that warrant attention by outpatient specialists and inpatient services. Fifth, we compare family-based h2 estimates, that were derived from a Swedish sample only, with h2-SNP results that were based on a variety of cohorts. Although these are all of the European descent, there might be heterogeneity in h2-SNP between cohorts, which makes the comparison with Swedish data less precise. We, therefore, examined heterogeneity in estimates of SCZ, as for this disorder a large Swedish sample contributed to the GWAS from which the h2-SNP was derived (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). Supplementary Table S7 of this study clearly shows that the h2-SNP based on the Swedish cohort equals h2-SNP estimates from other samples of similar size (i.e. Germany, UK), suggesting that the Swedish data are fairly comparable with other data from European descent.
Lastly, both designs assume random mating but a large-scale study in psychiatric populations (Nordsletten et al., Reference Nordsletten, Larsson, Crowley, Almqvist, Lichtenstein and Mataix-Cols2016) observed substantial non-random mating within and across disorders. A recent study, however, concluded that non-random mating has only a very modest effect on SNP-based heritability estimates in psychiatric traits (Peyrot et al., Reference Peyrot, Robinson, Penninx and Wray2016b).
Conclusion
In sum, this study presents a converging picture of the etiology of eight psychiatric disorders. SNP-based estimates were as expected lower but correlated with the family-based estimates. Additionally, the findings highlight large differences in genetic and environmental influences between psychiatric disorders. In contrast to ASD, ADHD, BIP, and SCZ, where genetic influences are most important, non-genetic influences play a large role in AD, AN, MDD, and OCD.
Conflict of interest disclosure
Dr H Larsson has served as a speaker for Eli-Lilly and Shire and has received research grants from Shire; all outside the submitted work. Dr P Lichtenstein has served as a speaker for Medice. Dr CM Bulik is grant recipient and consultant to Shire, and Consultant to Ironshore. Dr PF Sullivan is on the Scientific Advisory Board for Pfizer. Dr B Neale is on the Scientific Advisory Board for Deep Genomics.
Acknowledgements
Dr H Larsson acknowledges financial support through the Swedish Initiative for Research on Microdata in the Social And Medical Sciences (SIMSAM) framework Grant no. 340-2013-5867. Dr E Petterson acknowledges the Swedish Society for Medical Research. Dr CM Bulik acknowledges the Klarman Family Foundation, Wellcome Trust WT 088827/Z/09. The work of Dr Sullivan and the PGC MDD group was supported by NIMH U01 MH109528. Dr Walters and Dr Pardinas are supported by the UK Medical Research Council (Grants MR/L010305/1 and MR/L011794/1). Dr Børglum and the iPSYCH team acknowledge funding from The Lundbeck Foundation (grant no R102-A9118 and R155-2014-1724), the Stanley Medical Research Institute, an Advanced Grant from the European Research Council (project no: 294838), and grants from Aarhus University to the iSEQ and CIRRAU centers. Dr D Posthuma acknowledges funding from The Netherlands Organization for Scientific Research (NWO VICI 453-14-005).
The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0033291718002039.
Appendices
Sensitivity analyses
We conducted two sets of sensitivity analyses to examine whether the pre-specified age of the second sibling might have been too young. First, we conducted the analyses in the adult population only by including pairs in which the youngest sibling was 20 years old or older. Results are displayed in Table A1, and were very similar for all disorders except for OCD, for which the heritability estimate decreased from 0.38 to 0.24. However, due to a lack of maternal half-sibling cases, the standard error about this estimate was 0.20, indicating a lack of precision such that the two estimates did not differ significantly.
Second, we plotted the distributions of the age at first diagnosis for each disorder. We then re-ran the analyses but only included pairs where the younger sibling was at least as old as the median age of diagnosis. This way, the youngest siblings were more likely to have lived through the risk period of onset. Results are displayed in Table A2, and show that the estimates are remarkably similar to the original analyses (although the standard errors are larger).
In sum, the heritability estimates remained very similar regardless of whether we relied on our original age cutoffs, or on adults or the median age of onset.
Table A1. H2 of psychiatric diagnoses based on ICD 8, 9, and 10 in adults of >19 years

Table A2. H2 of psychiatric diagnoses based on ICD 8, 9 , and 10, with the younger sibling being at least as old as the median age of diagnosis







