Social media summary: Agta hunter–gatherer oral microbiomes are shaped by network structure and tradeoffs between sociality and disease spread
Introduction
Hominins have significantly diverged from other African apes regarding social behaviour and structure (Migliano & Vinicius, Reference Migliano and Vinicius2022). Compared with polygynous mating and male philopatric residence patterns typically found in chimpanzees, bonobos and gorillas, archaeological and ethnographic evidence points to a stepwise emergence of features such as pair bonding, multilocal residence, high mobility between residential camps and increased co-residence with unrelated individuals (Apicella et al., Reference Apicella, Marlowe, Fowler and Christakis2012; Hill et al., Reference Hill, Walker, Božičević, Eder, Headland, Hewlett and Wood2011). Such traits were the foundations of multilevel social structuring appearing in ancestral Homo sapiens and possibly earlier hominins. The niche of extant hunter–gatherers may offer a window into past human adaptations as it still exhibits some features prevalent before the advent of agriculture, such as a high-quality diet including meat and tubers, and multilevel sociality. Multilevel organisation results in interconnected social networks covering large areas and multiple residential camps (K. M. Smith et al., Reference Smith, Larroucau, Mabulla and Apicella2018), and in frequent interactions between individuals differing by sex, age and relatedness level. Interconnected networks may have accelerated the evolution of cultural innovations in humans compared with other apes (Derex & Boyd, Reference Derex and Boyd2016; Migliano et al., Reference Migliano, Battiston, Viguier, Page, Dyble, Schlaepfer and Vinicius2020). However, efficient networks may also facilitate the spread of infectious diseases (Page et al., Reference Page, Chaudhary, Viguier, Dyble, Thompson, Smith and Migliano2017), potentially affecting the structure and composition of hunter–gatherer microbiomes. Previous studies have investigated the role of diet, ecology and environment in hunter–gatherer oral, gut and milk microbiomes (Fragiadakis et al., Reference Fragiadakis, Smits, Sonnenburg, Van Treuren, Reid, Knight and Sonnenburg2019; Gomez et al., Reference Gomez, Petrzelkova, Burns, Yeoman, Amato, Vlckova and Blekhman2016; Meehan et al., Reference Meehan, Lackey, Hagen, Williams, Roulette, Helfrecht and McGuire2018; Moeller, Reference Moeller2017; Nasidze et al., Reference Nasidze, Li, Schroeder, Creasey, Li and Stoneking2011; Rampelli et al., Reference Rampelli, Schnorr, Consolandi, Turroni, Severgnini, Peano and Candela2015; Schnorr et al., Reference Schnorr, Candela, Rampelli, Centanni, Consolandi, Basaglia and Crittenden2014; Smits et al., Reference Smits, Leach, Sonnenburg, Gonzalez, Lichtman, Reid and Sonnenburg2017) and revealed higher oral microbiome diversity in hunter–gatherers than in farming populations (Lassalle et al., Reference Lassalle, Spagnoletti, Fumagalli, Shaw, Dyble, Walker and Balloux2018). However, they have not been able to isolate the contribution of high sociality and mobility to microbial transmission from other factors such as shared environments or diet. Although the more fluid and complex sociality of hunter–gatherers results in high levels of camp coresidence (Hill et al., Reference Hill, Walker, Božičević, Eder, Headland, Hewlett and Wood2011), cooperation and social interactions among unrelated individuals (Migliano et al., Reference Migliano, Page, Gómez-Gardeñes, Salali, Viguier, Dyble and Vinicius2017), its potential effects on microbiome transmission have not been fully determined. For this reason we present here a comprehensive investigation of the oral microbiome of Agta hunter–gatherers to analyse the specific effect of sociality and social network structure on its composition, supplementing an examination of the roles of environmental (diet) and biological (age, sex, host genotype) factors presented elsewhere (Dobon et al., Reference Dobon, Musciotto, Mira, Greenacre, Page, Dyble and Bertranpetit2021).
We obtained both oral microbiome sequences and high-resolution social network data from the same 138 Agta hunter–gatherers from the Philippines. We also collected oral microbiome data for 21 BaYaka hunter–gatherers from Congo Brazzavile, and 14 Palanan farmers neighbouring the Agta territory. We sequenced the 16S rRNA region and identified 6409 amplicon sequence variants (ASVs) (Callahan et al., Reference Callahan, McMurdie and Holmes2017), later reduced to 1980 ASVs (with at least 10 counts and present in at least two individuals), to detect fine-scale variation between individuals. We also collected data on proximity interactions and social networks using radio sensor technology that recorded close-range dyadic interactions every 2 minutes for 5–7 days (Migliano et al., Reference Migliano, Page, Gómez-Gardeñes, Salali, Viguier, Dyble and Vinicius2017, Reference Migliano, Battiston, Viguier, Page, Dyble, Schlaepfer and Vinicius2020) from four Agta camps, and from two longer multi-camp experiments (interactions recorded every hour for one month). Proximity data were supplemented with information on household composition, kinship and affinal relationships from all Agta individuals.
Our extensive dataset on oral microbiome composition and social interactions obtained from the same individuals allowed us to investigate in depth the possible effects of sociality on oral microbiome transmission and composition in Agta hunter–gatherers. Our aims were to investigate the roles of hunter–gatherer niche and geography on oral microbiome diversity in hunter–gatherers from two continents and a neighbouring farming population from the Philippines; determine which fraction of the Agta oral microbiome specifically responds to levels of social interaction; identify levels of pathogenicity of the oral microbiome transmitted through social contact; investigate potential tradeoffs between increased sociality and the spread of infectious disease; and verify potential tradeoffs at individual level by testing whether ‘hypersocial’ individuals also shared more bacteria. In the following, we provide evidence that the oral microbiome of extant hunter–gatherers was partially shaped by tradeoffs between extensive sociality and the spread of infectious disease.
Results
Hunter gatherer niches shape the oral microbiome
To investigate the contributions of lifestyle vs. environment to the hunter–gatherer oral microbiome, we compared the Agta (n = 138) with smaller samples of BaYaka hunter–gatherers from Congo Brazzaville (n = 21) and neighbouring Palanan farmers from the Philippines (n = 14) (see Methods). Both Agta (mean of 252.1 ± 90 ASVs per individual) and BaYaka (280.1 ± 83) exhibited significantly more ASVs than Palanan farmers (163.4 ± 34) (p < 0.0001; Figure 1a), and higher levels of ASV diversity as measured by Faith's Phylogenetic Diversity index (Figure 1b). Comparisons based on the total set of 6409 ASVs (controlling for differences in sample size through subsampling) revealed that the Agta shared more bacteria with African BaYaka (471.2 ± 33.9) than with neighbouring Palanan farmers (423.4 ± 23.8) (Figure 1c). Finally, Agta and BaYaka resampled groups showed respectively 651.5 ± 93.7 and 688.1 ± 61.9 exclusive ASVs, against only 285.1 ± 16.9 in Palanan farmers (Figure 1d). In summary, the two hunter–gatherer populations show higher microbiome diversity and uniqueness than Palanan farmers, consistent with findings that farming significantly reduced gut microbiome diversity (Schnorr et al., Reference Schnorr, Candela, Rampelli, Centanni, Consolandi, Basaglia and Crittenden2014). The results therefore demonstrate the precedence of niche over geography in shaping hunter–gatherer oral microbiomes.

Figure 1. Oral microbiome diversity in Agta hunter–gatherers, neighbouring Palanan farmers, and BaYaka hunter–gatherers. (a) Number of amplicon sequence variants (ASVs) in the Agta (n = 138), BaYaka (n = 21) and Palanan farmers (n = 14). (b) Oral microbiome diversity assessed by Faith's Phylogenetic Diversity index accounting for ASV phylogenetic distances (Agta = 17.52 ± 3.83; BaYaka = 18.45 ± 4.10; Palanan farmers = 12.62 ± 1.86). (c) Shared ASVs between populations, estimated by randomly sampling 10 individuals from each population (averaged over 100 permutations). (d) Exclusive ASVs per individual, estimated by randomly sampling 10 individuals from each population (100 permutations). Boxplot midlines represent medians, and box limits represent first and third quartiles (**** false discovery rate-adjusted p < 0.0001; *** p < 0.001; ** p < 0.01).
The social microbiome is a socially transmitted fraction of the oral microbiome
Primate social ‘pan-microbiomes’ were recently defined as the totality of microorganisms present in a host population or species (Sarkar et al., Reference Sarkar, Harty, Johnson, Moeller, Archie, Schell and Burnet2020), but this definition also includes microorganisms acquired owing to common diet or environment. Here we define the ‘social microbiome’ as the oral microbiome specifically transmitted through close-range social interactions (Migliano et al., Reference Migliano, Page, Gómez-Gardeñes, Salali, Viguier, Dyble and Vinicius2017). To identify the socially transmitted fraction of the Agta oral microbiome, we used the contact network derived from data collected by portable radio sensor devices and split all Agta dyadic social interactions into a strong (top 25% from the distribution of dyadic link weights) and a weak set (the remaining 75% links; see Methods). We then reduced our bacterial set to 1980 ASVs, or those present in at least two individuals and with an abundance of at least 10 counts per individual. We tested for differences in the proportion of each of the 1980 ASVs between the strong and weak sets. We identified 137 ASVs (7% of the Agta oral microbiome; see Supplementary Figure S1 and Supplementary Table S1) whose presence was significantly higher in the strong set, and therefore statistically associated with higher frequencies of social interactions. In the following we investigate the transmission patterns and composition of the hunter–gatherer social microbiome.
The hunter–gatherer social microbiome is predominantly pathogenic
Human sociality is associated with multiple fitness benefits, including increased reproductive success (Page et al., Reference Page, Chaudhary, Viguier, Dyble, Thompson, Smith and Migliano2017), reputation (Chaudhary et al., Reference Chaudhary, Salali, Thompson, Dyble, Page, Smith and Migliano2015), food sharing (Dyble et al., Reference Dyble, Thompson, Smith, Salali, Chaudhary, Page and Migliano2016), cooperation (Apicella et al., Reference Apicella, Marlowe, Fowler and Christakis2012; D. Smith et al., Reference Smith, Dyble, Thompson, Major, Page, Chaudhary and Mace2016) and cultural transmission (G. D. Salali et al., Reference Salali, Chaudhary, Thompson, Grace, van der Burgt, Dyble and Migliano2016), but may also facilitate pathogen transmission (Migliano et al., Reference Migliano, Page, Gómez-Gardeñes, Salali, Viguier, Dyble and Vinicius2017; Page et al., Reference Page, Chaudhary, Viguier, Dyble, Thompson, Smith and Migliano2017). In our dataset, from the 18 ASVs that could be classified at species level, 14 are socially transmitted, nine of which (64.3%) are typically pathogenic, and 10 (71.4%) are typically oral. In contrast, all four non-socially transmitted species were non-pathogenic and typically oral. We were able to classify 1886 of the 1980 ASVs at genus level, resulting in 36 socially (representing ASVs included in the social microbiome) and 62 non-socially transmitted genera (the remaining ones). Among the social genera, 61.8% were classified as typically or exclusively pathogenic (21 out of 34; two genera could not be classified), against only 16.4% among non-socially transmitted genera (10 out of 61; one genus could not be classified). We identified many socially transmitted genera either typically (Aggregatibacter, Capnocytophaga) or uniquely (Corynebacterium) associated with dental plaque formation, gingivitis and calculus, the full red complex of periodontal disease (Porphyromonas, Treponema and Tannerella), and other potential periodontal pathogens (Prevotella, Desulfobulbus and Fusobacterium) (Pérez-Chaparro et al., Reference Pérez-Chaparro, Gonçalves, Figueiredo, Faveri, Lobão, Tamashiro and Feres2014; Simón-Soro et al., Reference Simón-Soro, Guillen-Navarro and Mira2014; Simón-Soro & Mira, Reference Simón-Soro and Mira2015; Socransky et al., Reference Socransky, Haffajee, Cugini, Smith and Kent1998). It should be noticed that the classification of bacterial genera as pathogenic is not unequivocal in cases where some species within the genus are not pathogenic. Thus, once practical criteria were applied for assigning a genus to the pathogenic group (see Methods), the social microbiome was shown to exhibit a higher proportion of pathogenic genera than the non-socially transmitted fraction.
We also found pathogenic bacteria typical of the gut (Rickenellaceae), non-human environments (Tetragenococcus and Comamonas), respiratory tract (Staphylococcus, Moraxella and Streptococcus pneumoniae), urogenital and respiratory tracts (Mycoplasma), suggesting that their spread may be facilitated by oral transmission (Haq et al., Reference Haq, Battersby, Eastham and McKean2017; Jung et al., Reference Jung, Ehlers, Lombaard, Redelinghuys and Kock2017; Natsis & Cohen, Reference Natsis and Cohen2018). In summary, the predominantly pathogenic nature of the social microbiome suggests a tradeoff between benefits of hunter–gatherer sociality and costs associated with disease transmission.
Hunter–gatherer multilevel social structure shapes social microbiome sharing
Hunter–gatherer sociality is characterised by specific interaction channels not found in non-human apes, such as long-term pair bonding and households, extended families, friendships among unrelated individuals and frequent between-camp relocation. We estimated the effect of relatedness level, residence camp and friendships on the probability of sharing socially transmitted bacteria. First, we built a bacterial sharing network, where the weight of each Agta dyadic link is given by how many of the 137 social bacteria are shared by the two Agta individuals (rather than by the strength of its social bond, as in the social network). Next, we classified all dyadic links in this network based on kinship (mother–offspring, father–offspring, siblings, r = 0.5; other kin, r = 0.25 or r = 0.125; non-kin, r = 0.0625 or lower; spouses, friends, defined as non-kin at the top 25% distribution of social dyadic weights, and other non-kin) and residence (same or different camp, same or different household). We then compared the mean weight of each type of dyadic link in our bacterial sharing network with its mean weight in a sample of 1000 networks of the same size and topology, but where the dyadic classification was randomised (Figure 2 and Supplementary Tables S2 and S3).

Figure 2. Effect of kinship, friendship and residence on Agta dyadic bacterial sharing. Dyads were classified into kinship levels; same or different households; same or different camps; and between friends in the same or different camps. Dots show the z-score, or standardised ratio of mean link weight in real to randomised networks, in either social (orange) or non-socially transmitted bacteria (purple). Vertical red dashed line indicates a ratio of 1, or no difference between the number of shared bacteria in real and randomised networks. For socially transmitted bacteria, kinship, friendship and residence in the same household or camp are associated with significantly higher bacterial sharing than predicted from randomised networks of the same size and structure. In contrast, dyads from different camps or non-kin share significantly fewer bacteria than expected by chance; bacterial sharing in dyads from different households does not differ from randomised networks. For non-socially transmitted bacteria, the only dyadic categories significantly increasing bacterial sharing were siblings, spouses and dyads from the same household (all of which share the same close environment). See Supplementary Tables S2 and S3 for values on mean weights for real and randomised networks. (**** false discovery rate-adjusted p < 0.0001; *** p < 0.001; ** p < 0.01).
The results showed that some dyadic categories share significantly more socially transmitted bacteria than expected by chance. First, we observed higher bacterial sharing within the same household and camp, an expected consequence of the Agta multilevel social structure. Kinship effects were also clear, with the highest levels of social microbiome sharing found in mother–offspring pairs, followed by siblings known to interact daily in households and playgroups. High sharing between spouses also confirmed the importance of human pair bonding in microbial transmission. In addition, strong friendship links were also associated with increased bacterial sharing. Social bacterial sharing between friends in the same camp is as high as between close kin or within households. Friends in different camps also share a higher proportion of social bacteria than would be expected by chance, which is possibly a consequence of high between-camp mobility. In contrast, non-kin or individuals from different camps share fewer social bacteria than expected, further demonstrating the role of Agta friendships in the transmission of social bacteria across households and whole camps.
The same analysis performed instead on non-socially transmitted ASVs did not reveal significant effects on bacterial sharing from most dyadic categories, except for three types: spouses, siblings and same household. A possible explanation is that some non-socially transmitted bacteria may be shared not owing to interpersonal transmission but to a common environment and diet in the same household. For example, we have shown in a parallel study (Dobon et al., Reference Dobon, Musciotto, Mira, Greenacre, Page, Dyble and Bertranpetit2021) that the proportion of meat vs. rice in individual diets affects the composition of the Agta oral microbiome. Therefore, similar diets may explain the presence of the same ASVs within the individuals of a household irrespective of social interaction levels. However, the effects of sociality and shared diets seem to be independent. This is shown by the fact that socially transmitted bacteria are equally likely to be related or not to diet: 13 socially transmitted genera were also associated with diet (41 genera), whereas 23 were not (57 genera) (proportion test: χ2 = 0.44, p = 0.51). Our parallel study has also shown that host genotype correlates with the presence of certain ASVs. While high genetic relatedness may play a role in bacterial sharing within households, none of the ASVs associated with host genotypes were present in the social microbiome. Therefore, our analyses seem to distinguish between the effects of social contact from shared environment or genes within dyadic types. Overall, the results demonstrate the roles of mobility and the multiple interaction channels created by multilevel sociality in social microbiome sharing, similarly to those observed in cultural transmission (Dyble et al., Reference Dyble, Salali, Chaudhary, Page, Smith, Thompson and Migliano2015; Migliano et al., Reference Migliano, Page, Gómez-Gardeñes, Salali, Viguier, Dyble and Vinicius2017, Reference Migliano, Battiston, Viguier, Page, Dyble, Schlaepfer and Vinicius2020; Salali et al., Reference Salali, Chaudhary, Thompson, Grace, van der Burgt, Dyble and Migliano2016).
Frequency of social contact predicts social microbiome sharing
Although previous studies have investigated patterns of bacterial sharing in human groups, they have often been unable to comprehensively characterise transmission patterns owing to limited information on social contact (Brito et al., Reference Brito, Gurry, Zhao, Huang, Young, Shea and Alm2019). In order to obtain a full picture of individual contact and exposure levels, we built social networks based on proximity data from four camps and two multi-camp locations (Figure 3a, b). Overall, Agta social networks reveal a multilevel structure of households (mostly consisting of strong kin links) connected by a few strong links (mostly among unrelated friends) in each camp, and in the case of multi-camp groups, camps interconnected mostly owing to visits among friends. We also observed a close similarity in interactions within and between sexes and across age groups. This pattern creates multiple channels for social transmission of bacteria both within and between camps, between close kin and unrelated individuals, and finally across whole multi-camp structures.

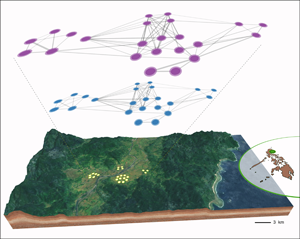
Figure 3. Characterisation of the social microbiome. (a) Recording networks of social interactions using radio sensor technology. (b) Reinforcement analysis predicts the probability of a link occurring in the bacterial sharing network (top layer, purple) from the weight of the same link in the social contact network (bottom layer, blue). Network nodes (circles) represent the same Agta individuals in the bacterial sharing and social contact network. Panel displays networks from multi-camp 1 (23 individuals). Map shows geographical location of four camps interconnected by frequent migration. (c) Probabilities of links in the Agta bacterial sharing network increase with their weights in the social contact network. Curves estimated by generalised additive modelling (binomial option). Data from four Agta camps and two multi-camps. (d) Eigenvector centralities in bacterial sharing and social contact networks. Linear regression based on pooled data from four Agta camps and two multi-camp structures. Virtually similar results were obtained by including camp either as a fixed factor in a multiple regression (with or without interactions), or as a random factor (on intercept and slope) in a mixed effects linear regression.
We applied reinforcement analysis (Battiston et al., Reference Battiston, Nicosia and Latora2014) (Figure 3b) to further assess whether the Agta social network predicted (or reinforces) bacterial sharing in each Agta camp. We calculated the conditional probability of each link between two individuals A and B in the bacterial network, provided the same weighted link was present in the social network (Supplementary Tables S4 and S5). For all four camps and two multi-camp structures, the results showed that the weight of a dyadic link in the social network significantly predicts the probability of the same link occurring in the bacterial sharing network (Figure 3c and Supplementary Figure S2). Specifically, a larger dyadic weight in the social network implies a higher probability that the same individuals also share at least one socially transmitted ASV. For example, for multi-camp 1, while weak social network links (with weights under 10 recorded social interactions) show a probability below 20% of sharing any socially transmitted bacteria, strong links (over 200 recorded contacts) are associated with a probability above 70%. Finally, we build another bacterial sharing network including all 1980 ASVs (93% of which are not socially transmitted). In this case, links in the social network cannot predict the presence of a bacterial sharing link, confirming that reinforcement specifically applies to the social microbiome. Overall, the results confirm that the Agta social microbiome is shaped by their social interactions.
Hypersocial individuals are supersharers
We also investigated whether more socially interactive individuals exhibited higher social microbiome diversity. We calculated eigenvector centralities for all individuals in the bacterial sharing network, resulting in a significant and positive slope in a regression on eigenvector centralities from the same individuals in the social network (b = 0.32, p = 0.0001, R 2 = 0.1, n = 138; Figure 3d). We also identified 16 individuals ranked at the top quartile of eigenvector centralities in both networks as potential microbial ‘superspreaders’ or ‘superacquirers’, that is, ‘supersharers’. They do not stem from a specific age (7–68 years) or sex (six males, 10 females), which is compatible with the egalitarian social structure of hunter–gatherers where individuals from any age or sex may be central to social networks.
Discussion
We have identified and characterised a socially transmitted fraction of the Agta hunter–gatherer oral microbiome. This fraction (7%) is surprisingly small, since in principle all 1980 identified ASVs could be orally transmitted between closely interacting people. Nonetheless, our results demonstrate a significant and independent role of social interactions on the transmission of oral microbiome, in addition to other factors such as shared environment (household and diet) and host characteristics (age, sex and genes) previously investigated in other hunter–gatherer populations (Fragiadakis et al., Reference Fragiadakis, Smits, Sonnenburg, Van Treuren, Reid, Knight and Sonnenburg2019; Gomez et al., Reference Gomez, Petrzelkova, Burns, Yeoman, Amato, Vlckova and Blekhman2016; Meehan et al., Reference Meehan, Lackey, Hagen, Williams, Roulette, Helfrecht and McGuire2018; Moeller, Reference Moeller2017; Nasidze et al., Reference Nasidze, Li, Schroeder, Creasey, Li and Stoneking2011; Schnorr, Reference Schnorr2015; Schnorr et al., Reference Schnorr, Candela, Rampelli, Centanni, Consolandi, Basaglia and Crittenden2014; Smits et al., Reference Smits, Leach, Sonnenburg, Gonzalez, Lichtman, Reid and Sonnenburg2017) and in the same Agta population (Dobon et al., Reference Dobon, Musciotto, Mira, Greenacre, Page, Dyble and Bertranpetit2021). The transmission of the 137 bacteria classified into the social microbiome seems to be facilitated by the extended sociality of hunter–gatherers and its various transmission channels, ranging from spouses to unrelated friends often residing in different camps. Together, reinforcement analysis, multiple channels of social interaction and supersharers show that social microbiome sharing is strongly shaped by hunter–gatherer multilevel sociality. From an evolutionary perspective, sociality has considerably changed from our closest ape relatives to ancestral humans, when adopting a hunter–gathering lifestyle meant exhibiting a higher frequency of social contact with unrelated individuals, larger networks of extended kin across large geographical regions, and more egalitarian interactions between and within sexes and across ages. Such changes may have affected patterns of pathogen transmission and affected the human microbiome, as observed in current hunter–gatherers.
As with our study of the Agta, future research should collect data on both social networks and social microbiomes from the same populations of non-human apes; such a dataset would provide a comparative basis for analysing of the role of social evolution on the human social microbiome. Hunter–gatherer social networks are efficient systems of cultural transmission, and specific channels organised around kinship, friendship and camp interconnectivity are central for the organisation of between-household cooperation, food sharing and social learning (Dyble et al., Reference Dyble, Thompson, Smith, Salali, Chaudhary, Page and Migliano2016; Salali et al., Reference Salali, Chaudhary, Thompson, Grace, van der Burgt, Dyble and Migliano2016; Smith et al., Reference Smith, Dyble, Thompson, Major, Page, Chaudhary and Mace2016). Agta mothers with higher social network centrality enjoy increased access to help and reproductive success (Page et al., Reference Page, Chaudhary, Viguier, Dyble, Thompson, Smith and Migliano2017), but our results have shown that efficient networks may also facilitate the spread of infectious diseases, and hence significantly affect the structure and composition of the Agta microbiome. Crucially the frequency of pathogenic bacteria is much higher in the socially than in the non-socially transmitted fraction of the Agta oral microbiome. Together with the association between hypersocial individuals and increased bacterial sharing, this suggests a tradeoff between potential fitness benefits and costs of increased pathogen transmission. We conclude that the predominantly pathogenic oral social microbiome we identified in Agta hunter–gatherers may be at least partially the outcome of a tradeoff between the advantages of multilevel sociality and the cost of infectious disease.
Methods
Ethnographic data collection
Agta demography
Ethnographic data collection took place over two seasons in April–June 2013 and February–October 2014. We censused 915 Agta individuals (54.7% male) across 20 camps. The four selected camps and two multi-camps in the current study were the ones providing social interaction data and saliva samples from almost all camp members. Accurate ages were estimated following relative aging protocols (Diekmann et al., Reference Diekmann, Smith, Gerbault, Dyble, Page, Chaudhary and Thomas2017). Relatedness (biological and affinal) was based on household genealogies. To resolve inconsistencies, we took either the genealogy from the most knowledgeable individual (i.e. mother over aunt) or the genealogy that reduced other inconsistencies (i.e. discarding 6 month interbirth intervals). Genealogies contained 2953 living and dead Agta. We used the R packages pedigree, kinship2 and igraph to measure consanguineous relatedness (r) (Dyble et al., Reference Dyble, Salali, Chaudhary, Page, Smith, Thompson and Migliano2015; Page et al., Reference Page, Myers, Dyble and Migliano2019). For comparative purposes, we obtained 14 saliva samples from neighbouring Palanan farmers, making sure that individuals were unrelated by directly asking.
BaYaka demography
Ethnographic data collection took place over two seasons in April–June 2013 and February–October 2014. We collected saliva samples from 21 individuals for microbiome analyses.
Ethics
This study was approved by UCL Ethics Committee (UCL Ethics code 3086/003) and carried out with permission from local government and community members. Informed consent was obtained from all participants, after group and individual explanation of research objectives in the indigenous language. A small compensation (usually a thermal bottle or cooking utensils) was given to each participant. The National Commission for Indigenous Peoples (NCIP) advised us that the process of Free Prior Informed Consent should be obtained from the community leaders, youth and elders under the supervision and validation of NCIP. This was done in 2017 with the presence of all community leaders, elders and youth representatives at the NCIP regional office, with the mediation of the regional officer and the NCIP Attorney. The validation process was approved unanimously by the community leaders and the NCIP, and validated the full 5 years of data collection.
Oral microbiome analysis
Microbial DNA extraction and 16S rRNA gene sequencing.
A total of 190 saliva samples were selected from Agta hunter–gatherers (n = 155) and Palanan farmers in the Philippines (n = 14), and BaYaka hunter–gatherers from the Congo (n = 21). Microbial DNA was extracted following the protocol for manual purification of DNA for Oragene⋅DNA/saliva samples. The 16S rRNA gene V3–V4 region was amplified by PCR with primers containing Illumina adapter overhang nucleotide sequences. All PCR products were validated through an agarose gel and purified with magnetic beads. Index PCR was then performed to create the final library also validated through an agarose gel. All samples were pooled together at equimolar proportions and the final pool was qPCR-quantified before MiSeq loading. Raw Illumina pair-end sequence data were demultiplexed and quality-filtered with QIIME 2 2019.1 (Bolyen et al., Reference Bolyen, Rideout, Dillon, Bokulich, Abnet, Al-Ghalith and Caporaso2019) and DADA2 (Callahan et al., Reference Callahan, McMurdie, Rosen, Han, Johnson and Holmes2016), which generates single nucleotide exact amplicon sequence variants (ASV or ESV). ASVs are biologically meaningful as they identify a specific sequence and allow for higher resolution than operational taxonomic units (OTUs) (Callahan et al., Reference Callahan, McMurdie and Holmes2017) or clusters of sequences above a similarity threshold, and thus an ASV is equivalent to a 100% similar OTU. Taxonomic information was assigned to ASVs using a naive Bayes taxonomy classifier against the SILVA database release 132 with a 99% identity sequence (Quast et al., Reference Quast, Pruesse, Yilmaz, Gerken, Schweer, Yarza and Glöckner2013).
Reads outside the kingdom Bacteria or assigned to mitochondria or chloroplasts were removed. Phylogenetic analyses aligned sequences with MAFFT (Katoh et al., Reference Katoh, Misawa, Kuma and Miyata2002) and generated a rooted phylogenetic tree with FastTree2 (Price et al., Reference Price, Dehal and Arkin2010) using default settings via QIIME 2. We generated an Alpha rarefaction curve with R package vegan to confirm that sample richness had been fully observed (Supplementary Figure S3).
Samples with extremely low number of reads (8000) were removed. This resulted in 6409 ASVs (later reduced to 1980 ASVs present in at least two individuals and with an abundance of at least 10 counts per individual) and 173 individuals: 138 Agta, 21 BaYaka and 14 Palanan farmers.
Identification of the Agta social microbiome
In our Agta sample, we first selected a set of strong social links (the top 25% of the weight distribution from each camp and multi-camp). For each ASV, we calculated the proportion of strong links ($f_{\rm A}^{\rm s}$![]() ) where a given ASV A was present. Next, we calculated the same proportion in the complementary set of 75% weak social links ($f_{\rm A}^{\rm w}$
) where a given ASV A was present. Next, we calculated the same proportion in the complementary set of 75% weak social links ($f_{\rm A}^{\rm w}$![]() ). We then computed for each ASV A the score
). We then computed for each ASV A the score
or normalised difference between the two proportions. This score can be paired with the Z-score

which quantifies the deviation from the null hypothesis that the two proportions are equal, with n s and n w as respectively the numbers of strong and weak links and f A as the proportion of total links that share ASV A. We then selected as affected by social interaction 137 ASVs with s > 0.5 and p < 0.05. The p-values were adjusted by false discovery rate.
We performed a sensitivity analysis to investigate the consequences of varying the threshold defining strong vs. weak social links in the Agta social network. Instead of 137 ASVs resulting from selecting dyads at top 25% of the weight distribution, we obtained:
top 45% – 166 ASVs;
top 35% – 156 ASVs;
top 25% – 137 ASVs;
top 15% – 138 ASVs;
top 5% – 99 ASVs.
The list shows that whether the strong set consists of a very reduced number of dyads with very strong weights (top 5%), or instead includes nearly half of the dyads (top 45%), the number of ASVs significantly responsive to social contact varies from 99 (5%) to 166 (8%), representing a small fraction of the total of 1980 ASVs found in the Agta. Therefore, setting the threshold at the top 25% did not affect our results and conclusions.
Agta social network data, construction and analysis
Portable radio sensor devices
Our wireless sensing devices store all between-device communications within a specified distance and have been described in detail elsewhere (Migliano et al., Reference Migliano, Page, Gómez-Gardeñes, Salali, Viguier, Dyble and Vinicius2017, Reference Migliano, Battiston, Viguier, Page, Dyble, Schlaepfer and Vinicius2020; Page et al., Reference Page, Chaudhary, Viguier, Dyble, Thompson, Smith and Migliano2017, Reference Page, Myers, Dyble and Migliano2019). We used the UCMote Mini (with a TinyOS operating system) sealed into wristbands or belts, labelled with a unique number and identified with coloured string to avoid accidental swaps. The devices require no grounded infrastructure and collect interactions even when individuals are away from camps. Individuals arriving at a camp after the start of data collection were given a device and the entry time was recorded, while those leaving a camp before the end of data collection had their exit time recorded. To prevent swaps individuals were checked twice daily, and device numbers were checked upon return. Any swaps were later corrected by reassigning data to the correct individuals.
Data were later downloaded via a PC side application in Java. Data were limited to 5 a.m. to 8 p.m. We ran raw data through a stringent data-processing system in Python to prevent data corruption. Data were matched to ID numbers and start-stop times of each sensor. The result was a matrix with the number of recorded beacons for all possible dyads and their weights.
For the camp-level experiment, all individuals from four camps wore sensors from 5 to 7 days. Each device sent a message every 2 minutes that contained its unique ID, a time stamp and the signal strength. Messages are stored by any other mote within a 3 m radius, a frequently used threshold (Brent et al., Reference Brent, Lehmann and Ramos-Fernández2011; Isella et al., Reference Isella, Stehlé, Barrat, Cattuto, Pinton and Van den Broeck2011). Detection of such close-range interactions is our proxy for social interactions. In previous studies, we have provided evidence that our sensor data capture interactions resulting from joint activities such as foraging, gathering, child care and socialising among others, through validation of sensor data by parallel observations obtained during camp scans (Migliano et al., Reference Migliano, Page, Gómez-Gardeñes, Salali, Viguier, Dyble and Vinicius2017; Page et al., Reference Page, Chaudhary, Viguier, Dyble, Thompson, Smith and Migliano2017). For the multi-camp experiment, adult individuals from two areas (consisting of seven and three camps respectively) wore sensors for 1 month.
Effect of dyad category on bacterial sharing
The bacterial sharing network was constructed by defining link weights as the number of social bacteria shared by two individuals. Dyads in the network were classified into: (i) levels of kinship (mother–offspring, father–offspring, siblings, r = 0.5; other kin, r = 0.25 or r = 0.125; non-kin, r = 0.0625; or lower, spouses, friends, defined as non-kin at the top 25% distribution of social dyadic weights, and other non-kin); and (ii) residence (same or different camp, same or different household). The mean weights were calculated for each dyadic category (Supplementary Table S2). Next we produced 1000 network randomisations based on a single-step ID swap between nodes. For example, if dyad 1 consisted of two spouses in the real network, randomisation preserved dyad 1 and its weight, but randomly replaced the two nodes (potentially changing the dyadic classification to siblings, friends, and so on). We calculated the mean weights for each dyadic category in the 1000 randomised networks, and then calculated one-sample t-tests with the mean weight in the real network as the test value. We repeated the analysis for non-social bacteria (Supplementary Table S3).
Reinforcement analysis
In a multilayer network, reinforcement analysis measures the overlap in links between different layers to quantify the probability of finding a link on a layer conditioned on the weight of the same link on another (Battiston et al., Reference Battiston, Nicosia and Latora2014). Reinforcement between two network layers α and α′, or P(α ′∨α), is defined as

where $a_{ij}^{[ {\alpha^{\prime}} ] }$![]() is the adjacency matrix of conditioned layer α′, and $w_{ij}^{[ \alpha ] }$
is the adjacency matrix of conditioned layer α′, and $w_{ij}^{[ \alpha ] }$![]() is the adjacency matrix of the conditioning layer α. We split the weight of social links into three tertiles, and computed equation (1) for each. We obtained increasing values of reinforcement from the lower to the higher tertile, providing evidence of an effect of social contact on bacterial sharing.
is the adjacency matrix of the conditioning layer α. We split the weight of social links into three tertiles, and computed equation (1) for each. We obtained increasing values of reinforcement from the lower to the higher tertile, providing evidence of an effect of social contact on bacterial sharing.
ASV classification
ASV diversity metrics
To distinguish between the effects of lifestyle and shared ecology on the microbiome, we compared the diversity of the oral microbiome of Agta hunter–gatherers with neighbouring Palanan farmers in the Philippines and BaYaka hunter–gatherers in Congo. Using the 6409 ASVs dataset we calculated the number of observed ASVs in each population with R package Phyloseq (version 1.30.0) and Faith's Phylogenetic Diversity index was calculated with R package picante using the generated rooted phylogenetic tree. To estimate the number of shared ASVs in the Agta, BaYaka and Palanan farmers, we sampled a random subset of 10 samples for each population without replacement, calculated the shared ASVs between the populations and repeated this procedure 100 times. Global differences between groups and pairwise comparisons were assessed by Kruskal–Wallis and Wilcoxon rank sum tests respectively and plotted by the R package ggpubr. Pairwise p-values were adjusted by false discovery rate.
Classification of oral bacteria as pathogens
ASVs were classified as oral pathogens if they have been reported as etiological agents of periodontitis (Pérez-Chaparro et al., Reference Pérez-Chaparro, Gonçalves, Figueiredo, Faveri, Lobão, Tamashiro and Feres2014; Socransky et al., Reference Socransky, Haffajee, Cugini, Smith and Kent1998) or dental caries (Simón-Soro et al., Reference Simón-Soro, Guillen-Navarro and Mira2014; Simón-Soro & Mira, Reference Simón-Soro and Mira2015). For gum disease pathogens, these included the classical ‘red’ and ‘orange’ complex of periodontal pathogens and the recent update (Pérez-Chaparro et al., Reference Pérez-Chaparro, Gonçalves, Figueiredo, Faveri, Lobão, Tamashiro and Feres2014) based on systematic review and metanalysis. For caries pathogens, the list of active microorganisms detected through metatranscriptomics of cavities was used. Common oral commensals potentially causing endocarditis or systemic infections in immunocompromised patients only were not considered pathogens. Bacteria reported as etiological agents of lower respiratory infections (e.g. pneumonia, whooping cough, bronchitis or sinusitis) and biofilm-mediated infections (e.g. lactational mastitis, medical implant biofilm infections, chronic lung infections, osteomyelitis or chronic wounds) were also considered pathogens, including organisms present in healthy carriers (Leung & Hon, Reference Leung and Hon2018; Natsis & Cohen, Reference Natsis and Cohen2018). Bacteria causing urinary tract infections or sexually transmitted diseases transiently found in the oral cavity were also considered pathogens (Jung et al., Reference Jung, Ehlers, Lombaard, Redelinghuys and Kock2017). Bacteria were classified as oral if detected in more than 10% of the population in oral samples according to the Human Oral Microbiome database. If a bacterial species or genus had been isolated from the oral cavity of another animal species, it was also classified as oral.
For assignment of bacteria as pathogenic or non-pathogenic, we used species-level ASVs, given the multiple cases where species from the same genus had a different assignment. If taxonomic classification of an ASV was only possible at the genus level, it was considered a pathogen if: (i) >90% of named species within the genus were pathogenic; or (ii) the genus included a major pathogenic species even if the remaining species were not classified as oral by the Human Oral Microbiome Database (Chen et al., Reference Chen, Yu, Izard, Baranova, Lakshmanan and Dewhirst2010). ASVs with a top hit to a sequence classified as ‘Oral taxa’ in databases but without a species assignment were discarded from the analysis. Cases where taxonomic classification of the ASV was only possible at the family level or higher were also discarded.
Acknowledgements
We are especially grateful to the Agta and BaYaka communities for their contribution to this study. We also thank the National Commission for Indigenous Peoples, Philippines, for their support.
Author contributions
ABM and JB conceived and designed the study. All authors collected and analysed data. ABM and LV wrote the article with assistance from all others.
Financial support
This project was funded by the Leverhulme Trust (grant RP2011-R 045 to ABM). JB received grant PID2019-110933GB-I00/AEI/10.13039/501100011033 from the Agencia Estatal de Investigación, Ministerio de Ciencia, Innovación y Universidades (Spain), support from the Secretaria d'Universitats i Recerca del Departament d'Economia i Coneixement de la Generalitat de Catalunya (grant GRC 2017 SGR 702), and support from the Agencia Estatal de Investigación (grant CEX2018-000792-M) to the ‘Unidad de Excelencia María de Maeztu’, Universitat Pompeu Fabra. AM is funded by grant RTI2018-102032-B-100 from Ministerio de Ciencia, Innovación y Universidades (Spain).
Conflicts of interest
The authors declare none.
Research transparency and reproducibility
16S amplicon data (EGAS00001005317) are deposited at the European Genome-phenome Archive, which is hosted at the European Bioinformatics Institute (EBI) and the CRG. Data at the individual level on age, kinship relationships, household composition, camp assignation and social contacts that support the findings of this study are available on request from the corresponding author (A.B.M.). The individual data are not publicly available owing to information that could compromise research participant privacy. All source code and data for visualisation are available at https://doi.org/10.5281/zenodo.6338840
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/ehs.2023.4