Rabies is a fatal viral zoonosis caused by viruses of the genus Lyssavirus in the family Rhabdoviridae. It is transmitted between mammals, including bats, primarily through bite inoculation of the rabies virus (RABV) present in the saliva of infected individuals [Reference Johnson1]. Members of the Lyssavirus genus constitute a single monophyletic clade, distinct from other rhabdoviruses. The genus consists of 11 genotypes (seven established genotypes and four newly described lyssaviruses from Eurasia). Genotype 1 (RABV, classical RABV) has worldwide distribution and at present is the only genotype to be isolated in the Americas (South, Central and North) that forms endemic cycles within terrestrial mammals and bats [Reference Kuzmin2].
Rabies occurs in two different epidemiological forms: urban rabies, with dogs and domestic animals as the principal reservoir and transmitter, and sylvatic rabies, with various wild species in the Carnivora and Chiroptera orders acting as reservoirs and transmitters. In Chile, dog rabies has been controlled, and since 1985 insectivorous bats have been identified as the country's main rabies reservoirs and infection source for sporadic cases of rabies in domestic animals [Reference Nuñez3, Reference Favi4].
At least four genera of insectivorous bats (Tadarida, Myotis, Histiotus, Lasiurus) are widely distributed in Chile. The role of these species as reservoirs hosts and transmitters supports the theory that diverse viral variants of rabies are circulating. Recent evidence suggests that all RABV variants affecting terrestrial carnivores may have originated from cross-species transmission events from long-term enzootic bat-associated variants. A molecular-clock model based on genetic divergence of RABV variants in bats of different species suggests that in North America, the divergence of extant bat-associated RABVs from a common ancestor took place between 1651 and 1660 c.e. The bat RABV variants found in Latin America in common vampire bats (Desmodus rotundus) and free-tailed bats (genus Tadarida, family Mollosidae) are the closest ones to the earliest common ancestor [Reference Calisher5].
The purpose of this study was to investigate which RABV variants were circulating in Chile in 2002–2008. During this period, 11 342 animals from areas around the country were submitted for rabies testing. Of this number, 653 insectivorous bats, one dog and one cat tested positive using the fluorescent antibody test (FAT). Applying the mouse inoculation test (MIT) [Reference Koprowsky, Kaplan and Koprowsky6], 612 samples were successfully isolated and then typed using a panel of eight monoclonal antibodies against the viral nucleoprotein (N-mAbs) provided by the Centers for Disease Control and Prevention (USA). The reaction patterns obtained with different mAbs for determining the antigenic variant have been described in a previous report [Reference Favi4].
Ninety-nine of the RABV isolates were selected for performing nucleotide sequence analyses. Of these, 66 were from T. brasiliensis bats (the most common species submitted for testing), 31 were from the remaining insectivorous bat species (L. cinereus, L. borealis, H. macrotus, M. chiloensis) and two were taken from domestic animals (dog and cat). All 99 were collected in Chile's central region (Fig. 1).

Fig. 1 [colour online]. Geographical distribution of sequenced rabies cases (Chile, 2002–2008).
Total RNA extraction was conducted using TRIzol (Invitrogen, USA) in accordance with the manufacturer's instructions. Complementary DNA (cDNA) was produced by reverse transcription–polymerase chain reaction using primers 10 g and 304 as described previously and a product of 320-bp of the nucleoprotein gene (1157–1476) was sequenced using the BigDye Terminator Cycle Sequence kit v3.1 (Applied Biosystems, USA) [Reference Yung, Fernández and Favi7]. Nucleotide sequences were analysed with an ABI PRISM 3130 genetic analyser (Applied Biosystems).
A phylogenetic tree was reconstructed for aligned nucleotide sequences by means of a neighbour-joining (NJ) analysis with 1000 bootstrap replicates using the MEGA 3 > 1 software tool [Reference Kumar, Tamura and Nei8]. Bootstrap resampling analysis of 1000 replicates was employed to estimate the reliability of the prediction tree. For the phylogenetic analysis, sequences from other countries in the Americas were included (GenBank accession numbers are given in Fig. 2). Two non-rabies lyssaviruses, European bat 1 (EBLV Genbank accession no. U22845) and Duvenhage virus (DUVV Genbank accession no. U22848) were used as outgroups [Reference Kissi, Tordo and Bourhy9].

Fig. 2. Phylogenetic relationships among 99 RABV isolates from Chile (GenBank accession nos. HQ385325–HQ385423) and rabies strains of bats from the Americas based on nucleotide homology of a 320-bp region of the nucleoprotein gene. Neighbour-joining tree with bootstrap values >50% obtained from 1000 resamplings are shown in the nodes. Tb, Tadarida brasiliensis; My, Myotis chiloensis; Hm, Histiotus macrotus; Lc, Lasiurus cinereus; Lb, Lasiurus borealis.
Reaction patterns using a panel of eight mAbs of 613 rabies isolates revealed six different antigenic variants in the Chilean bat species (Table 1). Of this total, 572 isolates were antigenic variant 4 (568 from T. brasiliensis bats, two from M. chiloensis bats and one each from a dog and a cat) and 18 were antigenic variant 6 (14 from L. cinereus bats, four from L. borealis bats). Eleven isolates from H. macrotus bats and one from T. brasiliensis bats were associated with an atypical antigenic variant described previously in Chile that is unrelated to any previously described reaction panel using a panel with eight N-mAbs [Reference Yung, Fernández and Favi7]. Five isolates from M. chiloensis bats were characterized as antigenic variant 3, two from M. chiloensis bats as variant 8, and four from T. brasiliensis bats as variant 9 (associated with T. brasiliensis mexicana).
Table 1. Antigenic typing with monoclonal antibodies (mAbs) of rabies isolates from Chile
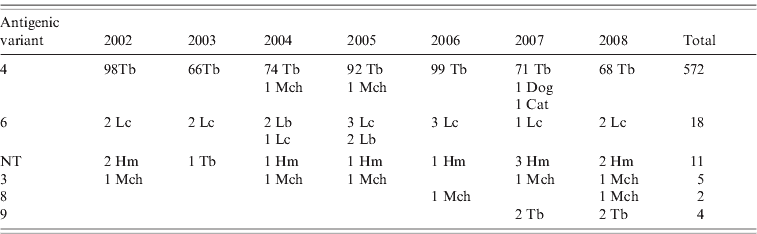
NT, Not typed; Tb, Tadarida brasiliensis; Lc, Lasiurus cinereus; Lb, Lasiurus borealis; Hm, Histiotus macrotus; Mch, Myotis chiloensis.
Rabies isolates are grouped according to patterns of reaction with eight N-mAbs.
Although it offers a rapid, simple and inexpensive means of typing for epidemiological studies, antigenic analysis with mAbs is lacking in precision. To obtain a more accurate determination of the diversity of the RABV in bat populations, partial sequencing and phylogenetic analyses of 99 Chilean RABV isolates were conducted. Four monophyletic clusters associated with four different bat species were thus identified, each one defined as a group of related sequences that share specific patterns of nucleotide variation and are associated with rabies maintained and transmitted by the same or some other bat species according to taxonomic identification of specimens (Fig. 2).
Cluster I contained 66 isolates obtained from 64 T. brasiliensis bats and two domestic animals (a dog and a cat), but due to the large number of isolates with 100% nucleotide similarity we took only representative sequences for the phylogenetics analyses. The overall average identity in these isolates was 95·9%. This variant is distantly related to the genetic variant circulating in the North American T. brasiliensis bat population but is very closely related to the genetic variants in Argentinean and Colombian bats. The RABV found in T. brasiliensis in Chile does not seem to be closely related to rabies in the same species in North America, where the RABV lineage found in T. brasiliensis is related primarily to the vampire viruses [Reference Velasco-Villa10]. Since RABV circulates in Chile in insectivorous bats only, it is not found in haematophagous bat species.
Cluster II was represented by isolates from six M. chiloensis bats (colonial and non-migratory) with an overall average identity of 95·5%. They were antigenically identified as variants 3 and 8 (Table 1), but in the genetic analysis they segregated into a different cluster associated with Argentinean Myotis bats.
Cluster III was composed of 10 isolates, nine from H. macrotus bats and one from a T. brasiliensis bat, with an overall average identity of 98·6%. These isolates clustered with viruses associated with H. macrotus in Argentina and a Histiotus-like bat found in Mexico [Reference Cisterna11]. Very little is known about the biology and distribution of this bat species, which may be found in other parts of the Americas in addition to Chile, Argentina and Mexico [Reference Velasco-Villa10].
Finally, Cluster IV was made up of 16 isolates, of which four were from L. borealis bats, 11 from L. cinereus bats and one from T. brasiliensis. The overall average identity was 99·5%. The Lasiurus genus is solitary and often described as a tree-dweller due to its roosting preference. It is also migratory and hence has a more southerly range during the winter. All three of these species share the same phylogenetic lineage as Lasiurus bats in North America. Some bat species seem able to maintain the same virus variant in geographically distant territories. The two T. brasiliensis cases observed in this cluster are probably spillovers of an endemic cycle maintained by Lasiurus sp. This spillover transmission mechanism may be due to the fact that solitary bat species such as Lasiurus spp. can develop furious rabies, in which case they may actively attack bats or other animals [Reference Kuzmin, Rupprech, Jackson and Wunner12].
One isolate (Mch-3171), obtained from a M. chiloensis bat and antigenically identified as variant 4, segregated into a different cluster, with an insectivorous bat from Colombia. It was more narrowly related to cluster II. However, given the lack of statistical support for its potential association with other RABVs so far reported, complete nucleoprotein sequences and a more comprehensive sampling encompassing RABV diversity in the region are needed to help identify whether it is a new variant or the reservoir host associated with it.
Although antigenic typing of RABV using mAbs may distinguish diverse variants of the virus, distinguishing different types within a variant may become difficult using this method, which could be more easily and accurately done with molecular characterization via nucleotide and amino-acid sequence determinations. These molecular analyses may help to unravel the precise genetic diversity of a RABV and the sequence characteristic of RABVs specifically associated with each host species. The first phylogenetic investigation into bat RABV using partial N gene sequencing established that there were distinct lineages of bat RABV associated with different bat species [Reference Smith13].
RABV is widespread in the Americas and genetic differentiation in RABVs is believed to have occurred in response to their association with particular host species [Reference Hughes, Orciari and Rupprecht14]. However, topography may play a less significant role in shaping the phylogeny of bat RABV than it potentially does for terrestrial mammal RABV [Reference Davis, Bourhy and Holmes15]. When a physical barrier is considerable (e.g. the Andes mountain range) genetic isolation may occur, as demonstrated by the separation of the Chilean strains from isolate samples obtained in other Latin American locations [Reference Kuzmin, Rupprech, Jackson and Wunner12].
In Chile, where long-term enzootic canine RABVs have not been detected since 1990, the disease is confined to the wild cycle mainly due to T. brasiliensis bats. Although no human rabies cases have been reported since 1996, rabies remains a public health risk in Chile and other parts of Latin America because of the frequency of contact between humans and bats. The coexistence of an abundant bat population with humans and their domestic animals in the urban centres of these countries poses a new challenge to the understanding of rabies epidemiology in metropolitan areas [Reference Favi16, Reference De Mattos17].
The approach adopted in this study enabled the identification of several rabies enzootics maintained independently by different species of insectivorous bat through transmission events involving bat-to-bat or bat-to-terrestrial species. The investigation also confirmed T. brasiliensis as the main RABV reservoir and the existence of compartmentalization in Chile in other bat species.
Finally, we note that studies of RABV characterization are a valuable asset in supporting epidemiological surveillance systems for the disease and selecting control strategies and monitoring programmes, which can have major impacts on both human health and ecosystems.
ACKNOWLEDGEMENTS
We thank the laboratory staff who assisted in this study for their excellent technical assistance and also thank Kenneth Rivkin for his valuable contribution in the translation of this paper. The authors are also grateful for funding provided by the Public Health Institute of Chile.
DECLARATION OF INTEREST
None.





