Shigella spp. is one of the most important causes of acute diarrhoea in developing countries and a significant cause of gastroenteritis in industrialized countries. It is most frequent in children, the elderly and the immunocompromised. In addition to diarrhoea, other symptoms include abdominal cramps, fever, nausea and vomiting. The infectious dose is very low (10–100 c.f.u.); it is usually spread from person to person through the faecal–oral route, and transmission is enhanced when hand-washing and overall hygiene are suboptimal although infection may occur from food or water sources [Reference Leclerc, Schwartzbrod and Dei-Cas1–Reference Chen, Chen and Chiu3]. Several outbreaks have been reported implicating a range of foods such as parsley, lettuce, cheese, oysters, etc., which are contaminated at origin or during food processing [4–Reference Kimura6]. Furthermore, acid foods when refrigerated support the survival of Shigella spp. and thus cause food poisoning [Reference Bagamboula, Uyttendaele and Debevere7].
In Argentina shigellosis is notifiable to the National Public Health Laboratories, as part of the activities of the National Laboratories Network for Surveillance of Diarrhoea and Foodborne Pathogens. This network is coordinated by the National Reference Laboratory (NRL) at the Instituto Nacional de Enfermedades Infecciosas – ANLIS ‘Carlos G. Malbrán’, where the isolates are submitted for further characterization, including serotyping and molecular subtyping. Most isolates submitted are associated with sporadic cases of diarrhoea and the most frequent species identified over the last 5 years are S. flexneri (72%) and S. sonnei (26%) which is consistent with frequencies of isolation in other developing countries. However, from January 2002 to February 2003, 981 isolates of Shigella spp. were submitted to the NRL; of these, 367 (37%) were S. sonnei and 614 (62%) were S. flexneri. The increase in S. sonnei was due mainly to an augmented number of isolates from three different areas of the country (Fig. 1) and this was confirmed by the local area public health authorities who notified that the number of stool cultures positive for S. sonnei had risen compared with those of previous years. An epidemiological investigation was carried out in each incident and further characterization of the clonal relationship between the isolates was performed at the NRL.

Fig. 1. Map of Argentina indicating the cities where the three incidents occurred.
Incident 1
An increase in the number of Shigella spp. isolates was noted in two laboratories in the city of Azul, Province of Buenos Aires between January and July 2002. A total of 82 faecal specimens were positive for S. sonnei, representing a rise of more than four-fold compared with the same time period in previous years. Infections affected both children and adults. Eleven S. sonnei isolates from patients with diarrhoea were available for further study.
Incident 2
In March and April 2002 an increase in the number of gastroenteritis cases was observed in the city of Neuquén, in the south of Argentina. As the region is defined as endemic for S. sonnei, the local authorities discounted the presence of an outbreak. Nevertheless, 14 isolates were submitted to the NRL for further characterization.
Incident 3
Epidemiological investigation showed that an outbreak of gastroenteritis which affected ∼2700 people occurred in the city of Salta, in the north of Argentina on 18 and 29 January 2003. The source of the outbreak was linked to the neighbourhood water supply but the organism was not recovered from the water samples. However, 17 patients had faecal samples culture-positive for S. sonnei and 10 of these isolates were available for typing.
S. sonnei is serologically homogeneous and other subtyping procedures, such as antimicrobial susceptibility patterns, plasmid profile analysis, PCR-based methods and pulsed-field gel electrophoresis (PFGE), have been used to discriminate among strains [Reference Navia8–Reference Liu11].
The 35 isolates from the suspected outbreak incidents were examined in this study, and compared against 21 S. sonnei strains from sporadic cases of shigellosis in different areas of Argentina obtained from the Enterobacteria National Culture Collection of the NRL. The species were identified by biochemical tests according to Edwards and Ewing [Reference Ewing12] and serotyped by slide agglutination using antisera provided by the Instituto Nacional de Producción de Biológicos – ANLIS ‘Carlos G. Malbrán’. Antimicrobial susceptibility testing was performed by Kirby–Bauer disc diffusion [Reference Bauer13] using the following antibiotics: trimethoprim–sulfamethoxazole (SXT, 1:25 μg), ampicillin (AMP, 10 μg), chloramphenicol (CHL, 30 μg), and ciprofloxacin (CIP, 5 μg). Interpretive standards were based on the Control Laboratory Standard Institute [14].
PFGE was carried out following the PulseNet standardized protocol for S. sonnei [15] using the restriction enzymes XbaI and BlnI. The PulseNet global standard strain Salmonella Braenderup H9812 was included every four lanes in each gel. The small-scale alkaline lysis protocol of Tolmasky and Crosa [Reference Tolmasky, Actis, Crosa and Hardy16] was applied with slight modifications for plasmid analysis. The Supercoiled DNA ladder (Invitrogen, Carlsbad, CA, USA) was used as molecular size marker.
PFGE patterns were assessed visually and by computerized image analysis using BioNumerics software, version 3.5 (Applied Maths, Sint-Martens-Latem, Belgium). The Dice similarity coefficient was applied to compare patterns and to establish genetic relationships between isolates. Clustering was based on the Unweighted Pair Group Method with Arithmetic averages (UPGMA) with a band position tolerance of 1·1% and optimization of 1·5%. The criteria suggested by Tenover et al. [Reference Tenover17] were applied for the designation of PFGE patterns and clusters. Thus, clusters of XbaI profiles were termed A–D and included patterns with up to six different bands and were located on the same branch of the dendrogram (Fig. 2, Table). Clusters of BlnI profiles were termed I–IV (Table). Plasmid profiles were analysed visually, patterns were considered different by the presence or absence of one or more bands and designated 1–10. Only clearly defined bands were considered in the analysis. Molecular size was assigned using Quantity One software, version 4.5.1 (Bio-Rad, Hercules, CA, USA).
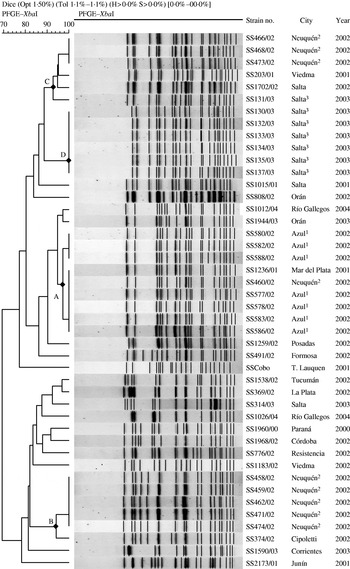
Fig. 2. Dendrogram obtained by cluster analysis of the PFGE patterns of S. sonnei isolates. Only the isolates corresponding to the outbreaks have a superior numerical indicator, for each one of the cities, as follows: Azul, 1; Neuquén, 2, and Salta, 3. DNA profiles of 21 sporadic strains are included for comparison.
Table. Characteristics of S. sonnei isolates from three suspected outbreaks in different regions of Argentina

AMP, ampicillin; SXT, Trimethoprim–sulfamethoxazole; CIP, ciprofloxacin.
The dendrogram of the similarity of the XbaI digests of outbreak and sporadic S. sonnei isolates by PFGE shows three major clusters of patterns at a cut-off of 90% that corresponded closely to the city of origin of the outbreak incident (Fig. 2). The 11 isolates from Incident 1 gave patterns that differed only by a single band of 70 kb which proved to be irreproducible on repeated testing and so were considered as a single clone (cluster A). Two sporadic isolates from different regions recovered in 2001 and 2002 were also indistinguishable from this clone. BlnI digests showed three highly similar patterns with only a two-band difference among the 11 isolates from Azul (Table) and supported the conclusion that all isolates from the outbreak were representative of a single clone. Thus, all the isolates from Azul were considered to be closely related and part of the outbreak. Isolates from Incident 2 in Neuquén fell into three different subtypes differing by 9–14 bands in XbaI DNA profile; nine were clustered together (cluster B), four were in cluster C and one grouped in cluster A with the isolates from Azul. Isolates representative of cluster B were also identified in Cipoletti, a city near Neuquén, and of cluster C in Viedma in the south and Salta in the north of the country. All, but one, of the isolates from Incident 3 in Salta were identical by XbaI profile, cluster D, and this clone was not identified in other areas. The remaining isolate differed by at least eight bands and clustered with pattern C (Fig. 2). Overall BlnI digests defined similar strain relationships to XbaI in PFGE and although two-band differences were observed between isolates of XbaI pattern B, this was insufficient to distinguish between the isolates by the Tenover criteria.
Ten different plasmid patterns were identified among the outbreak isolates ranging in size from 1·8 kb to ∼25 kb (Fig. 3). Isolates from Incident 1 were the most diverse in plasmid content with five different electrophoretic profiles among the 11 isolates; in Incident 2, eight of 14 isolates harboured the same plasmids and in Incident 3, nine of 10 isolates contained the same seven plasmids. Plasmid typing therefore correlated poorly with PFGE (Table).

Fig. 3. Line drawing representing the ten plasmid profiles of the S. sonnei outbreak related strains (labelled P1–P10), lane SDL: supercoiled DNA ladder.
Ampicillin resistance was detected in 14 isolates (all of those from the Salta outbreak) and was combined with resistance to cotrimoxazole in three isolates from Azul; all of the isolates from this outbreak were resistant to the latter agent. In contrast isolates from Neuquén were susceptible to all antibiotics assayed, except for one which was resistant to ampicillin and four isolates that were resistant to cotrimoxazole and were clustered in the XbaI–PFGE group C (Table).
Shigella spp. infections remain an important public health problem worldwide. It is therefore necessary for epidemiological surveillance to have highly discriminatory methods to trace the dissemination of strains both at the national and regional levels. In terms of population structure S. sonnei has for long been considered to be essentially a clonal subspecies of Escherichia coli as strains collected from widely distributed geographic sources were found to be identical by a number of molecular techniques [Reference Karaolis, Lan and Reeves18]. As in other clonal bacterial populations, evolutionary change occurs primarily through mutation as opposed to sexual recombination. This leads to a general lack of diversity in the population and it is therefore difficult to discern between strains at the molecular level for epidemiological studies.
In this study we applied the PulseNet standard PFGE protocol to investigate isolates of S. sonnei from three outbreaks in Argentina. The similarity of patterns obtained underlined the close genetic relationship among members of the serotype but the technique provided sufficient discrimination to indicate a single clone caused the gastroenteritis cases in two of the outbreaks. In the third centre three different lineages, which formed distinct clusters by similarity analysis, were circulating in Neuquén, in agreement with the epidemiological investigation which defined the area as endemic for Shigella spp. Nevertheless one of these clusters was predominant suggesting the possibility of an undetected outbreak situation. The finding of XbaI–PFGE pattern of the isolates from Azul in isolates from two other cities might point to the widespread circulation and distribution of this strain in different regions of Argentina. In contrast, the PFGE pattern exhibited by the isolates from Salta was not found in other strains previously isolated in the country.
Although plasmid profile analysis provided additional information as a molecular epidemiology tool for S. sonnei, there was not always a high correlation with the PFGE results. However, the plasmid patterns showed better correlation with the epidemiological data when the isolates were recovered in short time periods. Thus, the isolates from the Salta outbreak, recovered over 11 days exhibited a nearly identical plasmid profile with excellent correlation with PFGE. On the other hand, Neuquén isolates recovered over 2 months showed six different plasmid patterns; one of which included isolates in two distinct PFGE clusters suggesting that the same plasmids had been acquired by distinct clones. Lastly, in Azul, where the isolates were recovered over 7 months, five plasmid patterns were identified in isolates highly similar by PFGE.
It is unlikely that sequence-based typing based on housekeeping genes such as multilocus sequence typing would give increased discrimination for epidemiological typing of S. sonnei as an earlier study indexing variation in 12 housekeeping genes using multilocus enzyme electrophoresis found high homogeneity among 20 strains from widely geographical sources [Reference Ochman19]. PFGE probably offers the highest discriminatory power over other methods because it gives an overview of the general chromosomal architecture of the species rather than defining allelic variation in single gene loci which owing to the lack of recombination and low mutational rates are highly conserved in S. sonnei [Reference Karaolis, Lan and Reeves18]. In this context a global PFGE study of S. sonnei strains using the standard protocol would be beneficial in accurately defining the extent of variation present in DNA macrorestriction profiles of unrelated strains and establishing the band difference criteria applicable to this species.
Multiple antimicrobial resistance has been increasing in the last years among strains of Shigella spp., as reported in different regions worldwide [Reference Navia8, 20–Reference Alcoba22]. Among the S. sonnei isolates studied here, only those from Azul showed resistance to more than one antimicrobial agent. All the isolates from Neuquén that were resistant to cotrimoxazole showed the same PFGE patterns and also exhibited the same plasmid profile. We were therefore unable to ascertain whether the resistance was horizontally or vertically transmitted among these isolates. Furthermore, resistance to this drug was also found in the isolates from Azul, which displayed different PFGE and plasmid profiles.
Although at least two outbreak strains were identified, no sources for the infection could be confirmed by the laboratory. In Azul, screens for S. sonnei in different food samples yielded negative results but the duration of the outbreak over several months might have indicated predominantly person-to-person transmission. S. sonnei was not isolated from the neighbourhood water supply even though faecal contamination of water was demonstrated and the epidemiological investigation pointed to this as the source of infection. Isolation of Shigella spp. from food and water is often difficult [Reference Wachmuth, Morris and Doyle23] and similar experiences have been described in other outbreak settings [2,Reference Alcoba22]. There is, therefore, a need for the development of highly sensitive methods for the detection of these organisms, usually found in low numbers, in food and water.
In conclusion, PFGE grouped together epidemiologically related isolates of S. sonnei from two outbreaks in Argentina and distinguished them from non-related strains. Some of the genetic clusters identified included isolates from very distant locations, although no epidemiological link was established between them. This could be due to travel of individuals or consumption of contaminated food distributed throughout the country or alternatively the wide dissemination of these clones could have occurred long before the beginning of this study. Plasmid analysis provided some useful information as a preliminary screen only for isolates recovered over short time periods. The PFGE patterns of these clones have been incorporated into the national S. sonnei PFGE database and this information will provide the starting point for the active surveillance of shigellosis and for outbreak investigations in Argentina and Latin America. This database will be shared with other members of PulseNet International.
ACKNOWLEDGEMENTS
We thank the ‘Servicio Sueros y Antígenos’, from the Instituto Nacional de Producción de Biológicos, ANLIS ‘Carlos G. Malbrán’ for providing the antisera for Shigella serotyping. Also, the collaboration of Maria Inés Caffer, Angela Salve, Marcela Panagopulo, and Ofelia Martinez, from the INEI-ANLIS ‘Carlos G. Malbrán’ is gratefully acknowledged. We especially thank Dr Swaminathan, from CDC and Dr Enrique Pérez, from INPPAZ-PAHO/WHO, for their constant support and invaluable help in the implementation of PulseNet Latin America.
DECLARATION OF INTEREST
None.






