Fetal programming, or the adaptive responses of the fetus to a variety of environmental factors, and the consequences of mismatch between prenatal and post-natal environments are now known to shape development and metabolism, potentially contributing to adult-onset disease (Barker & Osmond, Reference Barker and Osmond1986; Barker et al., Reference Barker, Winter, Osmond, Margetts and Simmonds1989; Barker et al., Reference Barker, Hales, Fall, Osmond, Phipps and Clark1993a; Reference Barker, Osmond, Simmonds and Wield1993b). LBW is defined as birth weight lower than 2,500 grams in singleton births (World Health Organization, 1992). Studies have linked LBW to childhood mortality (McCormick, Reference McCormick1985), morbidity (Wu et al., Reference Wu, Xing, Fuentes-Afflick, Danielson, Smith and Gilbert2011), and childhood asthma (Brooks et al., Reference Brooks, Byrd, Weitzman, Auinger and McBride2001). LBW is also associated with disorders progressing into adulthood, such as metabolic syndrome (Fagerberg et al., Reference Fagerberg, Bondjers and Nilsson2004), type 2 diabetes (Johansson et al., Reference Johansson, Iliadou, Bergvall, de Faire, Kramer, Pawitan and Cnattingius2008), cardiovascular diseases (Leeson et al., Reference Leeson, Kattenhorn, Morley, Lucas and Deanfield2001), respiratory diseases (Walter et al., Reference Walter, Ehlenbach, Hotchkin, Chien and Koepsell2009), and depression (Thompson et al., Reference Thompson, Syddall, Rodin, Osmond and Barker2001).
Two important factors that influence birth weight are the length of the gestation period and the prenatal growth rate. Other modifiable factors exist in utero, such as maternal smoking during pregnancy (Ward et al., Reference Ward, Lewis and Coleman2007) and maternal health and nutrition (e.g., caloric intake; Kramer, Reference Kramer1987a, Reference Kramer1987b). Several genetic variants have been associated with birth weight (Freathy et al., Reference Freathy, Mook-Kanamori, Sovio, Prokopenko, Timpson, Berry and McCarthy2010; Horikoshi et al., Reference Horikoshi, Yaghootkar, Mook-Kanamori, Sovio, Taal, Hennig and Freathy2013); however, these show only a modest genetic contribution to the total variance in birth weight (Barker, Reference Barker2004; Battaglia & Lubchenco, Reference Battaglia and Lubchenco1967; Freathy et al., Reference Freathy, Mook-Kanamori, Sovio, Prokopenko, Timpson, Berry and McCarthy2010; Heijmans et al., Reference Heijmans, Tobi, Stein, Putter, Blauw, Susser, Slagboom and Lumey2008; Horikoshi et al., Reference Horikoshi, Yaghootkar, Mook-Kanamori, Sovio, Taal, Hennig and Freathy2013; Jarvelin et al., Reference Jarvelin, Sovio, King, Lauren, Xu, McCarthy and Elliott2004; McIntire et al., Reference McIntire, Bloom, Casey and Leveno1999). Heritability estimates for birth weight from twin and family studies have given highly variable results, ranging from 15% to 72% (Baird et al., Reference Baird, Osmond, MacGregor, Snieder, Hales and Phillips2001; Clausson et al., Reference Clausson, Lichtenstein and Cnattingius2000; Gielen et al., Reference Gielen, Lindsey, Derom, Smeets, Souren, Paulussen, Derom and Nijhuis2008; Lunde et al., Reference Lunde, Melve, Gjessing, Skjaerven and Irgens2007; Magnus, Reference Magnus1984a, Reference Magnus1984b; Magnus et al., Reference Magnus, Gjessing, Skrondal and Skjaerven2001; Mook-Kanamori et al., Reference Mook-Kanamori, van Beijsterveldt, Steegers, Aulchenko, Raat, Hofman, Eilers and Jaddoe2012). Given the high relevance of LBW to health throughout the life course, the molecular links between birth weight and age-related disease has attracted much research.
LBW may be associated with epigenetic variation, and recent evidence suggests that maternal nutrition intake is a crucial factor determining the extent of this impact (Amarasekera et al., Reference Amarasekera, Noakes, Strickland, Saffery, Martino and Prescott2014; Dominguez-Salas et al., Reference Dominguez-Salas, Moore, Baker, Bergen, Cox, Dyer and Hennig2014; Fernandez-Twinn & Ozanne, Reference Fernandez-Twinn and Ozanne2010). A number of environmental factors, including smoking, stress, hypoxia, a diet high in methyl-donor nutrients, such as folic acid, folate, and vitamin B during pregnancy, may alter gene expression during early embryonic development when epigenomes are becoming established. For example, DNA methylation levels at the IGF2 gene were reported to be significantly lower in adults who were in utero during the Dutch Hunger Winter Famine (1944–1945) when compared to same-sex siblings in utero unexposed to famine (Heijmans et al., Reference Heijmans, Tobi, Stein, Putter, Blauw, Susser, Slagboom and Lumey2008). Other candidate gene studies have also reported birth weight related methylation differences in imprinted genes that play a role in fetal growth, such as IGF2 and H19 (Hoyo et al., Reference Hoyo, Fortner, Murtha, Schildkraut, Soubry, Demark-Wahnefried and Murphy2012; Steegers–Theunissen et al., Reference Steegers-Theunissen, Obermann-Borst, Kremer, Lindemans, Siebel, Steegers, Slagboom and Heijmans2009) and in non-imprinted genes such as the glucocorticoid receptor NR3C1 (Filiberto et al., Reference Filiberto, Maccani, Koestler, Wilhelm-Benartzi, Avissar-Whiting, Banister, Gagne and Marsit2011; Mulligan et al., Reference Mulligan, D'Errico, Stees and Hughes2012). More recently, epigenome-wide association studies (EWAS) have been performed for birth weight using the Infinium HumanMethylation27 BeadChip (Infinium 27K; Adkins et al., Reference Adkins, Tylavsky and Krushkal2012; Banister et al., Reference Banister, Koestler, Maccani, Padbury, Houseman and Marsit2011; Fryer et al., Reference Fryer, Emes, Ismail, Haworth, Mein, Carroll and Farrell2011; Straughen et al., Reference Straughen, Sipahi, Uddin, Misra and Misra2015) and the Infinium HumanMethylation450 BeadChip (Infinium 450K; Engel et al., Reference Engel, Joubert, Wu, Olshan, Haberg, Ueland and London2014; Simpkin et al., Reference Simpkin, Suderman, Gaunt, Lyttleton, McArdle, Ring and Relton2015) methylation platforms (Illumina Inc, San Diego, CA). The results show that methylation at a few CpG sites appears to be associated with birth weight, but the magnitude of methylation differences tends to be small. For example, in one study a 10% difference in (untransformed) DNA methylation levels at the majority of birth weight associated CpG sites was reported to correspond to covariate-adjusted birth weight differences of 200 grams or less, with few exceptions (Engel et al., Reference Engel, Joubert, Wu, Olshan, Haberg, Ueland and London2014). One longitudinal study in singletons found that birth weight associated methylation changes are restricted to individuals of a younger age, and that these effects did not persist into older age (Simpkin et al., Reference Simpkin, Suderman, Gaunt, Lyttleton, McArdle, Ring and Relton2015). However, all of these studies were performed in unrelated subjects, where maternal environment and exposures are difficult to adequately account for.
Here, we explored DNA methylation differences in MZ twins discordant for birth weight. The discordant MZ twin study design can control for many potential epigenetic confounders; specifically, differences in genetic background, shared early life environmental exposures, age, sex, and cohort effects. Although MZ twins do not necessarily share the same in utero environment, they are more closely matched than unrelated individuals, and they offer a unique opportunity to study the link between early life factors and adult life health. Birth weight discordance in twins may arise through mechanisms not experienced by singletons (Lopriore et al., Reference Lopriore, Nagel, Vandenbussche and Walther2003), but shared results linking birth weight to cardiovascular health are identified in both twins and singletons (McNeill et al., Reference McNeill, Tuya and Smith2004). A recent longitudinal study in twins from birth to 18 months explored genome-wide DNA methylation patterns in buccal epithelium (Martino et al., Reference Martino, Loke, Gordon, Ollikainen, Cruickshank, Saffery and Craig2013). The authors observed that the extent of weight discordance in twins was greater at birth compared to at 18 months, which they termed a convergence in discordance. In contrast, the extent of methylation discordance in twins over time exhibited a twin-pair-specific pattern, where some twin pairs showed epigenetic convergence over time, some pairs showed epigenetic divergence of time, and other pairs remained equally discordant at birth and at 18 months. The observation that both weight and epigenetic profiles can exhibit convergence in twin pair discordance over time suggests that epigenetic studies of birth weight in twins may yield fruitful for uncovering epigenetic signatures of birth weight, but that the observed effects should be explored further in twin samples obtained at birth, ideally in a longitudinal study design.
Several studies in twins have explored epigenetic and gene expression profiles in relation to birth weight or maternal factors strongly associated with birth weight (Gordon et al., Reference Gordon, Joo, Andronikos, Ollikainen, Wallace, Umstad and Craig2011, Reference Gordon, Joo, Powell, Ollikainen, Novakovic, Li and Saffery2012; Loke et al., Reference Loke, Novakovic, Ollikainen, Wallace, Umstad, Permezel and Craig2013; Ollikainen et al., Reference Ollikainen, Smith, Joo, Ng, Andronikos, Novakovic, Abdul and Craig2010; Souren et al., Reference Souren, Lutsik, Gasparoni, Tierling, Gries, Riemenschneider and Walter2013; Tan et al., Reference Tan, Frost, Heijmans, von Bornemann Hjelmborg, Tobi, Christensen and Christiansen2014). To date, three EWAS have focused on MZ twins discordant for birth weight. The first profiled DNA methylation using the Infinium 27K array in 14 birth weight discordant MZ twin pairs (Gordon et al., Reference Gordon, Joo, Powell, Ollikainen, Novakovic, Li and Saffery2012). One differential methylation signal was identified in the APOLD1 gene, which has previously been linked to metabolic phenotypes. The effects appeared to be specific to human umbilical vascular endothelial cells, and significant changes were not observed in placental tissue or cord blood mononuclear cells. The second study used Infinium 450K to compare the methylation levels in buccal samples from 17 monochorionic MZ adult twins (Souren et al., Reference Souren, Lutsik, Gasparoni, Tierling, Gries, Riemenschneider and Walter2013). By comparing the methylation differences between the heavy and light co-twins and adjusting the methylation levels with leukocyte subtypes, the authors identified 3,153 birth weight differential methylation sites at p < .01 and 45 of these showed absolute methylation differences greater than 5%, but none of these changes reached genome-wide significance. The third more recent study in blood Infinium 450K profiles from 150 adult MZ twin pairs also found no genome-wide associations with birth weight in the overall sample, but analysis of 28 extremely birth weight discordant MZ twins pairs identified three significant differentially methylated sites, where the effects appeared to be restricted to specific age groups; that is, the intra-pair differential methylation effect either increased or decreased with age (Tan et al., Reference Tan, Frost, Heijmans, von Bornemann Hjelmborg, Tobi, Christensen and Christiansen2014). In this study, we explored the hypothesis that birth weight differences in MZ twins are reflected in the whole blood epigenome and persist into adulthood.
Materials and Methods
Study Population, Phenotype, and Covariate Data
Discovery sample: TwinsUK cohort
We used phenotypes and DNA methylation data generated on individuals from the TwinsUK cohort. The cohort was established in 1992 and recruited MZ and DZ same-sex twins (Moayyeri et al., Reference Moayyeri, Hammond, Valdes and Spector2013). The majority of participants are healthy female Caucasians (age range from 16 to 98 years old). There are over 13,000 twin participants across the United Kingdom and many have returned for multiple visits. We selected 71 MZ twin pairs from the cohort who were extremely discordant for birth weight (see discordance criteria below) for DNA methylation profiling.
Each subject's blood sample was collected during a clinical visit, along with a clinical questionnaire that included information on self-reported birth weight, medical history, and measures for height, weight, and BMI. The age at DNA extraction was taken into account as a covariate since methylation levels can change over time (Levesque et al., Reference Levesque, Casey, Szyf, Ismaylova, Ly, Verner and Booij2014; Martino et al., Reference Martino, Loke, Gordon, Ollikainen, Cruickshank, Saffery and Craig2013). Additional covariates, such as smoking status and alcohol consumption, were collected by self-reported questionnaires. Subjects were categorized as non-smokers, ex-smokers, and current smokers, and alcohol consumption data was summarized as grams/day intake. Participants were asked for their weekly average amount of alcohol intake (e.g., wine, beer, spirits, and liqueurs) and alcohol consumption data were then summarized as units per week, and then converted to grams/day (one unit of alcohol in the UK is defined as 7.9 grams; Brennan et al., Reference Brennan, Meng, Holmes, Hill-McManus and Meier2014). Previous studies have reported that the composition of white blood cells (WBC) can influence DNA methylation levels (Houseman et al., Reference Houseman, Accomando, Koestler, Christensen, Marsit and Kelsey2012). We obtained six cell count estimates, including: CD8+ T cells, CD4+ T cells, natural killer cells, CD19+ B cells, CD14+ monocytes, and granulocytes using previous methods (Houseman et al., Reference Houseman, Accomando, Koestler, Christensen, Marsit and Kelsey2012).
Birth weight discordance for each twin pair was calculated as the birth weight ratio (BWratio; Martino et al., Reference Martino, Loke, Gordon, Ollikainen, Cruickshank, Saffery and Craig2013; Tan et al., Reference Tan, Frost, Heijmans, von Bornemann Hjelmborg, Tobi, Christensen and Christiansen2014) as follows:
We considered birth weight discordant MZ twins where BW ratio >20%.
Replication samples 1 and 2: Danish Twin Registry (DTR)
The Danish Twin Registry was established in the 1950s and has collected information on over 88,000 twin pairs born in Denmark. Here, we used birth weight and whole blood DNA methylation Infinium 450K data from 27 old birth weight discordant MZ twin pairs with BWratio >20% (replication sample 1, DTR old), and from 29 young birth weight discordant MZ twin pairs with BWratio >20% (replication sample 2, DTR young). The phenotype and methylation data in these samples have previously been described (Frost et al., Reference Frost, Petersen, Brixen, Beck-Nielsen, Holst, Christiansen, Hojlund and Christensen2012; Tan et al., Reference Tan, Frost, Heijmans, von Bornemann Hjelmborg, Tobi, Christensen and Christiansen2014). Birth weight information in the DTR old sample was derived from midwife reports, and information in the DTR young sample was obtained from the Danish Birth Record Registry. The methylation data were first normalized using subset quantile within-array normalization method (Maksimovic et al., Reference Maksimovic, Gordon and Oshlack2012) and then normalized to a normal distribution using a logit transformation. The methylation residuals were taken after adjusting for covariates such as age, sex, and blood cell composition. The resulting intra-pair methylation differences then correlated to the BWratio using Spearman's correlation test.
Replication sample 3: Netherlands Twin Register (NTR)
The Netherlands Twin Register was established in 1987 and the NTR survey studies and the NTR BioBank project are described elsewhere (Boomsma et al., Reference Boomsma, Vink, van Beijsterveldt, de Geus, Beem, Mulder and van Baal2002, Reference Boomsma, de Geus, Vink, Stubbe, Distel, Hottenga and Willemsen2006; Willemsen et al., Reference Willemsen, de Geus, Bartels, van Beijsterveldt, Brooks, Estourgie-van Burk and Boomsma2010, Reference Willemsen, Vink, Abdellaoui, den Braber, van Beek, Draisma and Boomsma2013). Genome-wide methylation in blood was profiled using the Infinium 450K platform (Van Dongen et al., 2015). Data on birth weight were obtained from self-report by the twins themselves or their parents. Data collected across multiple surveys and projects were combined and consistency across family members and time was checked. When multiple data points differed by less than 200 grams, the average was taken, and larger differences data were excluded. Here, we analyzed birth weight and blood DNA methylation data that were available for 89 birth weight discordant MZ twin pairs, with BWratio >20%. The methylation data were first subject to functional normalization (Fortin et al., Reference Fortin, Labbe, Lemire, Zanke, Hudson, Fertig, Greenwood and Hansen2014) and then transformed to follow the normal distribution. A linear regression was used to fit the data to take into account the following covariates: sex, age, methylation array row, methylation plate, smoking status (non-smokers, ex-smokers, and current smokers), neutrophil percentage, monocyte percentage, and eosinophil percentage. The resulting residuals were used to calculate intra-pair methylation differences, and these were correlated with BWratio using Spearman's correlation.
DNA Methylation
In the discovery sample, DNA was extracted from whole blood and bisulfite converted prior to DNA methylation analysis using the DNeasy kit (Qiagen, Inc.). Bisulfite modification was performed using 96 well EZ DNA methylation kit (Zymo Research) with 750 ng of DNA sample. Methylation levels were detected using the Infinium 450K and the intensity images captured by GenomeStudio (2010.3) Methylation module (1.8.5) software. Details of the method are described elsewhere for a subset of our sample (Tsaprouni et al., Reference Tsaprouni, Yang, Bell, Dick, Kanoni, Nisbet and Deloukas2014). The beta mixture quantile dilation (BMIQ) method was performed to correct for the technical issues caused by the two Illumina probe types (Teschendorff et al., Reference Teschendorff, Marabita, Lechner, Bartlett, Tegner, Gomez-Cabrero and Beck2013). DNA methylation probes that mapped to multiple locations (with exact sequence match and within up to two base pair mismatches) to the reference sequence were removed. Probes for which more than 1% subjects had detection p value >.05 were also removed. A background signal correction by removing the signal detected from a negative control samples were applied to the methylation data using R package ‘Minfi’ (Aryee et al., Reference Aryee, Jaffe, Corrada-Bravo, Ladd-Acosta, Feinberg, Hansen and Irizarry2014). Subjects with over 5% missing probes were removed. We only considered autosomal probes in the analysis because the discovery sample consisted of females only, while the replication samples included both males and females. The final methylation data contained 442,307 probes with no missing values. Because beta values are overall not normally distributed, probe data was transformed using quantile normalization to a standard normal distribution.
Methylation Analysis
The association between DNA methylation and birth weight was investigated for all probes in the discovery dataset of 71 birth-weight discordant MZ twins. Principal component analysis (PCA) was applied to the methylation data to identify batch effects by correlating each potential covariate with the first two PCs, which together explained 23.7% of the variance. A linear mixed effect regression model was applied to the available methylation data, and methylation residuals were obtained after adjusting for covariates. The covariates in the model included age, WBC count estimates, smoking status (non-smokers, ex-smokers, and current smokers), alcohol consumption (grams/day), methylation plate, and position on the plate as fixed effects, and family and zygosity as random effects.
To explore the association between birth weight discordance and DNA methylation in the MZ twin samples, we first considered birth weight as a continuous trait. Intra-pair methylation differences were calculated by subtracting the methylation levels from the larger twin minus their smaller co-twin. The methylation differences were then correlated with the birth weight discordance to identify differential methylation sites, and the Spearman's correlation coefficients and p values were reported at each CpG site. In addition, we also assessed whether significant DNA methylation differences exist between the larger and smaller co-twin in the sample, irrespective of the exact birth weight values. In this case, twins were assigned to two groups (based on larger and smaller birth weight) and methylation levels were compared within-pairs using the Wilcoxon signed-rank test. The MZ twin continuous analyses were also pursued at the top-ranked probe in replication samples from the Netherlands and from Denmark.
Meta-Analysis of Twin Datasets
We analyzed the top-ranked signal in the discovery and replication datasets using a meta-analysis of the correlation coefficients, performed using the R packages ‘metacor’ (Laliberté, Reference Laliberté2011) and ‘Mac’ (Re & Hoyt, Reference Re and Hoyt2012). We first measured the diversity index of the studies (I2 ) to detect the homogeneity among the studies using the ‘Mac’ package (Higgins & Thompson, Reference Higgins and Thompson2002) and then performed the meta-analysis based on either the random-effects or the fixed-effects model (Schulze, Reference Schulze2004).
Results
Demographic Characteristics of the Twin Samples
The descriptive characteristics of the discovery cohort and the replication cohorts are shown in Table 1. The discovery sample for epigenome-wide analysis consisted of 71 birth weight discordant MZ female twin pairs (mean age 55.4 years, age range: 34.2–77.7 years) without severe poor health conditions, including cancer and type 2 diabetes, at the age of DNA sample collection. Birth weight discordance was defined as BWratio greater than 20% (see Methods), and the discovery mean BWratio was 35.0% (range: 20.4–71.3%), corresponding to a mean absolute birth weight difference of 943 g (±562 g). Subjects with a smaller birth weight were significantly shorter in adulthood height (p = 4.88 × 10−9), as observed in previous studies, but no significant association was found with BMI (p = .947), and marginal significance was obtained with adult weight (p = .036). No significant association was observed with age at blood sampling and BWratio. We calculated the intra-pair correlation in genome-wide DNA methylation Infinium 450K profiles in the discovery sample of 71 MZ pairs, and observed high correlation (correlation coefficient = 0.997, range: 0.987–0.999), which is consistent with previous studies (Bell et al., Reference Bell, Tsai, Yang, Pidsley, Nisbet, Glass and Deloukas2012; Souren et al., Reference Souren, Lutsik, Gasparoni, Tierling, Gries, Riemenschneider and Walter2013; van Dongen et al., Reference van Dongen, Ehli, Slieker, Bartels, Weber, Davies and Boomsma2014) and suggests minimal within-pair methylation differences.
TABLE 1 Characteristics of Four Birth Weight Discordant MZ Twin Samples

N = number of MZ twin pairs; Age = DNA extraction age; BWdiff = absolute birth weight difference within twin pairs (g); BWratio = birth weight ratio within twin pairs (%); NTR = Netherlands Twin Registry; DTR = Danish Twin Register.
We pursued replication of the top-ranked LBW-associated DNA methylation signal found in the discovery sample in three additional independent samples of birth weight discordant MZ twin pairs from Denmark (replication samples 1 [DTR old] and 2 [DTR young]) and the Netherlands (replication sample 3 [NTR]; Table 1). In each case, we only selected birth weight discordant MZ twin pairs with BWratio greater than 20%. However, the extent of birth-weight discordance in the replication samples was attenuated compared to the discovery cohort. The replication samples consisted of adult MZ twins either younger (NTR, DTR young) or older (DTR old) than the mean age of the discovery cohort, and contained individuals of both sexes.
Epigenome-Wide Analysis of Birth Weight Discordant Twins
Epigenome-wide analyses in the discovery sample of 71 MZ twin pairs compared methylation differences within pairs to BWratio using Spearman's correlation (Figure 1A). One CpG site passed genome-wide significance based on FDR of 0.05. The signal was obtained at CpG site cg12562232 (correlation coefficient = 0.603, 95% CI: 0.430–0.719, p = 2.62 × 10−8), which is located in intron 2 of the IGF1R gene on chromosome 15. At this site there was a positive correlation between intra-pair methylation differences and BWratio (Figure 2A). We also checked the other 128 CpG sites within the promoter or gene body of the IGF1R gene for association with birth weight. Although none of these CpG sites reached genome-wide significance, one signal located in the IGF1R gene body (cg23091737), and within 5 kb from cg12562232, surpassed nominal significance in the same direction of association. Aside from IGF1R, the next top-ranked signals were obtained at cg12049992 in the FAM38B gene (correlation coefficient = -0.519, p = 3.49 × 10−6), cg12508856 in the KIF13B gene (correlation coefficient = -0.510, p = 5.52 × 10−6), cg12391576 in the HLA-DPA1 gene (correlation coefficient = 0.508, p = 6.07 × 10−6), and cg26313699 in the OR1G1 gene (correlation coefficient = -0.507, p = 6.29 × 10−6) but the results were not genome-wide significant (FDR = 0.57).

FIGURE 1 Manhattan plots of birth weight EWAS in 71 MZ discordant twins using (A) BW as a continuous trait, and (B) BW as a categorical trait. The red point above the 5% FDR line (grey dashed line) is the birth weight differential methylation signal at cg12562232 in the IGF1R gene.
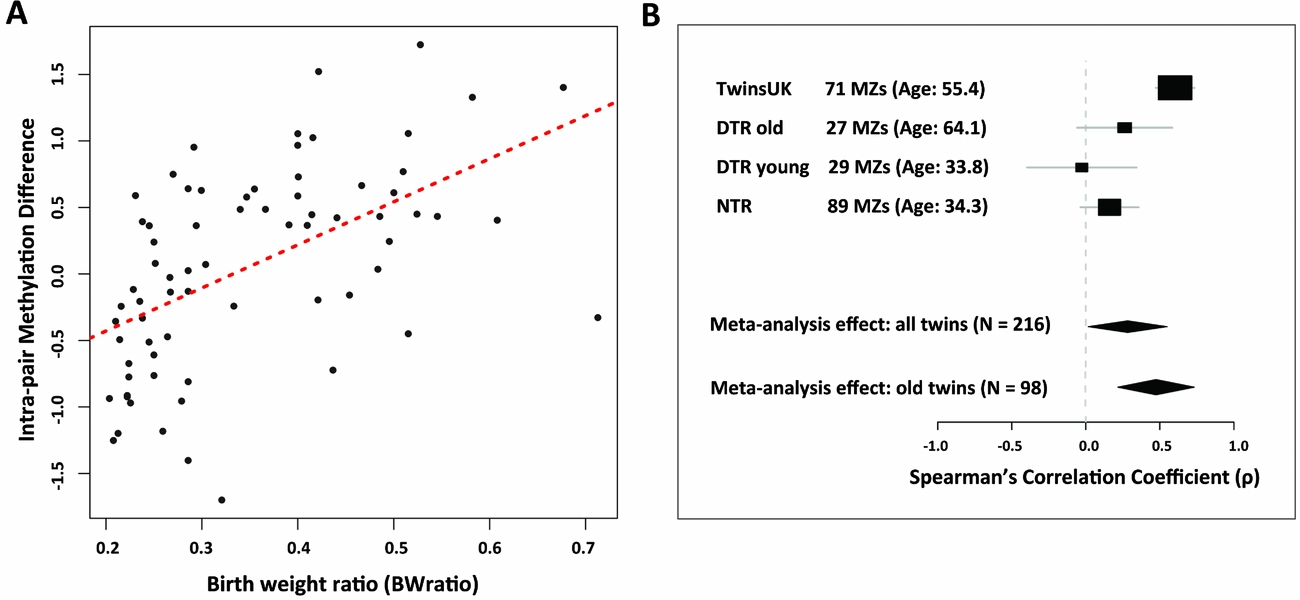
FIGURE 2 Birth weight differential methylation signal in IGF1R. (A) The correlation between the covariate-adjusted intra-pair methylation residual differences and the birth weight ratio in 71 birth weight discordant MZ pairs from TwinsUK. Each dot represents a single pair. (B) Forest plot of the meta-analysis results at the IGF1R gene in four birth weight discordant MZ twin samples from the United Kingdom (TwinsUK), Denmark (DTR old and DTR young), and the Netherlands (NTR). Random-effects meta-analysis estimates are shown for the results encompassing all four samples, as well as the two older twin samples from TwinsUK and DTR old.
We also explored intra-pair methylation differences between the larger twin and their smaller co-twin, without considering the exact value of the birth weight discordance. The epigenome-wide analysis in this category was performed using non-parametric paired tests and no CpG sites reached genome-wide significance (Figure 1B). The top-ranked signal in these analyses was obtained at CpG site cg24296900 located 1.5 kb from the transcription start site of the TUBA1 C gene (p = 3.27 × 10−7).
Replication of the IGF1R Signal
In order to try and replicate the association between birth weight discordance and IGF1R methylation levels, we performed a similar analysis in three independent MZ twin samples from the Danish (N = 29 and N = 27 MZ pairs) and the Netherlands (N = 89 MZ pairs) twin registries. DTR samples were stratified by age at recruitment, and therefore we performed the correlation test separately in the young (N = 29) and old (N = 27) twins. In contrast to the TwinsUK sample, the replication samples differed in mean age, contained male subjects, and were overall less discordant for birth weight (Table 1). Compared to the discovery correlation coefficient of 0.603 between IGF1R methylation and birth weight ratio in TwinsUK, a similar but not significant correlation was found in the DTR old (correlation coefficient = 0.263, p = 0.186) and NTR (correlation coefficient = 0.161, p = 0.132) samples (Table 2). The correlation was near zero in the DTR young sample (correlation coefficient = -0.027, p = 0.888; Table 2). We considered that a sex-specific effect may be present at this probe and performed sex-specific analysis. A similar positive correlation was found in both male and female subsets, suggesting that there is no sex-specific effect on the probe.
TABLE 2 Random-Effects Meta-Analysis Results in the IGF1R Gene (cg12562232)
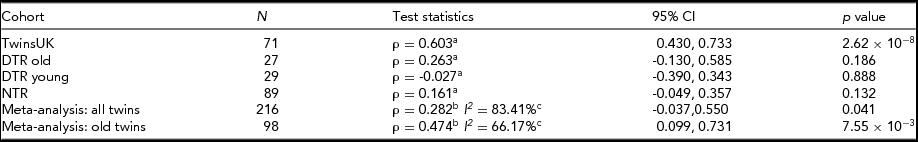
aSpearman's correlation coefficient.
b mean correlation coefficient.
c Heterogeneity Index.
Meta-Analysis at IGF1R
We next performed a meta-analysis of the correlation coefficients between IGF1R methylation and birth weight ratio across the four birth weight discordant MZ samples (Table 2). We observed heterogeneity among the four samples (I2 = 83.4%), and therefore performed a random-effects meta-analysis. The results showed a significant positive association (correlation coefficient = 0.282, 95% CI: -0.037, 0.550, p = 0.041) between IGF1R methylation and birth weight ratio across the four studies; in total, 216 MZ twin pairs (Figure 2B).
Compared to the TwinsUK and the DTR old samples, the DTR young and the NTR samples are much younger. The majority (84.3%) of the MZ pairs in the DTR young sample are younger than 45 years old, whereas 84.6% of the TwinsUK pairs are older than 45 years old. Considering the age differences across the samples and the high level of heterogeneity observed across the four studies, we also performed meta-analysis across studies stratified by age. A meta-analysis of the two older age samples (N = 98 TwinsUK and DTR old twin pairs) did not have strong evidence for heterogeneity (I2 = 66.2%), and the results showed a much stronger association in the random-effects meta-analysis (correlation coefficient = 0.474, 95% CI: 0.099, 0.731, p = .008) (Figure 2B, Table 2). Given the weak evidence for heterogeneity in the two older age samples, we therefore also performed a fixed-effects meta-analysis of these two samples and observed strong evidence for association (correlation coefficient = 0.5527, 95% CI: 0.415, 0.690, p = 4.59 × 10−12). These findings suggest that the observed association between birth weight and DNA methylation at the IGF1R gene is more pronounced at an older age.
Discussion
We performed epigenome-wide association analyses in whole blood samples of extreme birth weight discordant MZ twin pairs. We identified one significant differential methylation site in the IGF1R gene in the TwinsUK cohort. We observed a similar direction of effect in two independent MZ twin replication samples, and the combined meta-analysis of four twin samples showed a nominally significant association between IGF1R methylation and birth weight ratio (random-effects meta-analysis p = .041). Analyses stratified by age group indicated that the meta-analysis effect was accentuated in older twins. Although DNA methylation profiles at this CpG site have not been associated with age, to our knowledge (Horvath, Reference Horvath2013; Marttila et al., Reference Marttila, Kananen, Hayrynen, Jylhava, Nevalainen, Hervonen and Hurme2015; Steegenga et al., Reference Steegenga, Boekschoten, Lute, Hooiveld, de Groot, Morris and Muller2014), and are not associated with age in the TwinsUK sample, the mean age and age range of the four meta-analysis samples is very different and the results strongly suggest that the birth weight association effect is more pronounced at older age groups (TwinsUK and DTR old). Twin pairs from the TwinsUK cohort were also more discordant for the birth weight compared to all replication samples, and this might be an explanation for the attenuated effects observed in the replication samples, where the methylation differences in less discordant twins are modest. Our results are consistent with previous birth weight studies of the adult methylome in discordant MZ twins (Souren et al., Reference Souren, Lutsik, Gasparoni, Tierling, Gries, Riemenschneider and Walter2013; Tan et al., Reference Tan, Frost, Heijmans, von Bornemann Hjelmborg, Tobi, Christensen and Christiansen2014), that is, that there is no strong association between specific methylation signal and birth weight discordance, but moderate changes are observed.
One factor influencing the results is power to detect modest effect sizes in our study. In our previous power estimation study, a mean methylation difference of 12% was required to reach 80% power with a sample size of 71 MZ pairs (Tsai & Bell, Reference Tsai and Bell2015). However, the observed MZ twin pair difference in IGF1R related to birth weight was much smaller, and corresponded to up to approximately a 3% change in the unadjusted raw DNA methylation levels. The methylation profiles reported in older MZ twins (Fraga et al., Reference Fraga, Ballestar, Paz, Ropero, Setien, Ballestar and Esteller2005) and in heritability studies (Bell et al., Reference Bell, Tsai, Yang, Pidsley, Nisbet, Glass and Deloukas2012; Gordon et al., Reference Gordon, Joo, Powell, Ollikainen, Novakovic, Li and Saffery2012; Kaminsky et al., Reference Kaminsky, Tang, Wang, Ptak, Oh, Wong and Petronis2009) show that MZ twins tend to have very similar methylation patterns over the genome. This may be one reason explaining why none of the epigenetic studies in twins has identified large effects associated with birth weight.
Significant variation in the neonatal methylome occurs during early development and with changes in the intrauterine environment (Gordon et al., Reference Gordon, Joo, Powell, Ollikainen, Novakovic, Li and Saffery2012; Levesque et al., Reference Levesque, Casey, Szyf, Ismaylova, Ly, Verner and Booij2014; Martino et al., Reference Martino, Loke, Gordon, Ollikainen, Cruickshank, Saffery and Craig2013). Therefore, a number of studies have focused on birth weight associated methylation changes in candidate imprinted genes in both newborns and adults, and found associations in the H19 and IGF2 genes (Heijmans et al., Reference Heijmans, Tobi, Stein, Putter, Blauw, Susser, Slagboom and Lumey2008; Hoyo et al., Reference Hoyo, Fortner, Murtha, Schildkraut, Soubry, Demark-Wahnefried and Murphy2012; Murphy et al., Reference Murphy, Ibanez, Hattersley and Tost2012). In addition to birth weight associated changes at candidate genes, epigenome-wide analyses have also explored the top-ranked birth weight associated signals and have identified these to be enriched for genes related to early cell and embryonic development, growth, immune system, and inflammatory response (Adkins et al., Reference Adkins, Tylavsky and Krushkal2012; Banister et al., Reference Banister, Koestler, Maccani, Padbury, Houseman and Marsit2011; Engel et al., Reference Engel, Joubert, Wu, Olshan, Haberg, Ueland and London2014; Fryer et al., Reference Fryer, Emes, Ismail, Haworth, Mein, Carroll and Farrell2011; Gordon et al., Reference Gordon, Joo, Andronikos, Ollikainen, Wallace, Umstad and Craig2011; Reference Gordon, Joo, Powell, Ollikainen, Novakovic, Li and Saffery2012; Simpkin et al., Reference Simpkin, Suderman, Gaunt, Lyttleton, McArdle, Ring and Relton2015; Straughen et al., Reference Straughen, Sipahi, Uddin, Misra and Misra2015).
We observed a significant association between lower birth weight ratio and a lower methylation level at IGF1R in twins who were free from severe diseases, for example, cancer and type 2 diabetes, at the time of DNA methylation sampling, The insulin-like growth factor 1 receptor gene encodes a trans-membrane receptor that mediates the effects of IGF1 and where the receptor is activated in part by IGF2. Both IGF1 and IGF2 play a critical role in growth and have been linked to birth weight in genetic, epigenetic, and gene expression studies (Adkins et al., Reference Adkins, Somes, Morrison, Hill, Watson, Magann and Krushkal2010; Heijmans et al., Reference Heijmans, Tobi, Stein, Putter, Blauw, Susser, Slagboom and Lumey2008; Koutsaki et al., Reference Koutsaki, Sifakis, Zaravinos, Koutroulakis, Koukoura and Spandidos2011; McMinn et al., Reference McMinn, Wei, Schupf, Cusmai, Johnson, Smith, Weksberg and Tycko2006; Ong et al., Reference Ong, Kratzsch, Kiess, Costello, Scott and Dunger2000; Straughen et al., Reference Straughen, Sipahi, Uddin, Misra and Misra2015). Twin studies of IGF1 have found that intra-pair birth weight differences were significantly correlated with intra-pair IGF1 expression differences (Canpolat et al., Reference Canpolat, Cekmez, Sarici, Korkmaz and Yurdakok2011) and blood IGF1 concentrations (Westwood et al., Reference Westwood, Gibson, Sooranna, Ward, Neilson and Bajoria2001). Furthermore, mutations in IGF1R can lead to abnormalities in the function of IGF1 receptors that may slow down intrauterine and subsequent growth (Abuzzahab et al., Reference Abuzzahab, Schneider, Goddard, Grigorescu, Lautier and Keller2003). Previous work has also explored differential methylation in the IGF1R gene with birth weight. One study compared 34 maternal impaired glucose tolerance (IGT) to 106 normal glucose tolerance placentas and found that IGF1R methylation levels were significantly lower in placentas exposed to IGT (Desgagne et al., Reference Desgagne, Hivert, St-Pierre, Guay, Baillargeon, Perron and Bouchard2014). While it is suspected that IGF1R may have dysregulated fetal methylation levels related to birth weight, the cause-effect relationship has not been established. In our results, birth weight discordance is associated with methylation at a CpG site in intron 2 of IGF1R, which overlapped a weak ENCODE DNase signal in a number of cell types, including primary Th1T cells, suggesting potential regulatory effects. Recent preliminary work has also reported that gene expression levels of IGF1R were significantly lower in placentas from healthy mothers of term newborns who were of small gestational age (Lazo-de-la-Vega-Monroy et al., Reference Lazo-de-la-Vega-Monroy, González-Domínguez, Daza-Benítez and Barbosa-Sabanero2015), although this association was not observed in chorionic villus samples (Demetriou et al., Reference Demetriou, Abu-Amero, Thomas, Ishida, Aggarwal, Al-Olabi and Moore2014). Our findings therefore complement these results, and suggest that some of these effects may also be observed in the fetus and could be epigenetically mediated.
The pathways leading to LBW and birth weight discordance in twins may arise through mechanisms that are not experienced by singletons (Lopriore et al., Reference Lopriore, Nagel, Vandenbussche and Walther2003). Generally, singletons and twins develop at a similar rate until the 30th week of gestation, after which point uterine restriction is a major contributing factor to twin birth weight (Cleary-Goldman & D'Alton, Reference Cleary-Goldman and D'Alton2008). A lighter MZ twin has a greater likelihood of being genuinely growth restricted compared to their co-twin (Torche & Echevarria, Reference Torche and Echevarria2011). Birth weight differences in twins are thought to originate from multiple factors, including differences in fetal access to nutrition, which can be affected by the position of fetus in utero, as well as the umbilical cord. Twin-to-twin transfusion syndrome (TTTS) is another potential cause of the birth weight discordance in twins. The prevalence of TTTS is relatively low on average (1–3 per 10,000 births), but is greater in monochorionic twins, and can account for up to 17% of the total perinatal morality in twins, and 50% of death in MCDA twins (Steinberg et al., Reference Steinberg, Hurley, Desmedt and Beischer1990; Lewi et al., Reference Lewi, Jani, Blickstein, Huber, Gucciardo, Van Mieghem and Deprest2008).
There are several limitations in our study. First, birth weight or body weight is a complex phenotype, and the exact contribution of genetic and epigenetic variation to it, and extent of its shared effects with late-onset diseases, remains largely unknown. We used MZ twin pairs in the study to minimize the impact of genetic variation. Second, the sample size was moderate, and given the observed small effect at the top-ranked signal, larger samples will be needed to reach sufficient power. Third, information on the chronicity of the twin pairs was not available, and previous studies have reported that MZ MC (monochorionic: twins that share a single placenta) twins have a more imbalanced nutrient supply than the MZ DC (dichorionic: twins with two separate placentas) twins (Derom et al., Reference Derom, Vlietinck, Thiery, Leroy, Fryns and Derom2006). Fourth, the birth weight information in the TwinsUK cohort was self-reported, which could potentially influence the intra-pair discordance and effect estimates. Lastly, although our study identifies a significant association between LBW and a DNA methylation change in the IGF1R gene, which is highly relevant to growth, we cannot rule out the potential effects of reverse causation or confounding on the findings. Although the twins included in our sample are self-reported to be free from severe disease, this is not necessarily a reflection of their entire post-natal health and environmental exposures in early life and over the life course. Longitudinal epigenetic studies in twins would be the ideal study design to assess the epigenetic signature of birth weight discordance and its health impacts over the life course.
Conclusion
We found one genome-wide significant differential methylation signal in the IGF1R gene in discordant identical twin pairs, assessed many years after birth, which was confirmed in a meta-analysis of four independent twin samples. Our findings in adult twins suggest that methylation changes associated with prenatal conditions such as LBW may persist over time, but that the observed effects tend to be relatively modest.
Acknowledgments
We would like to thank all twins and family members in the TwinsUK cohort, the NTR, and the DTR.
TwinsUK: Support for this work was obtained from the European Research Council (ERC 250157) and in part from the TwinsUK resource, which is funded by the Wellcome Trust; the European Community's Seventh Framework Programme (FP7/2007–2013); and the National Institute for Health Research (NIHR) BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy's and St Thomas’ NHS Foundation Trust and King's College London.
NTR: We would like to thank the funding agencies for their support, including the Netherlands Organization for Scientific Research (NWO 900-562-137, 904-61-090, 985-10-002, 904-61-193, 56-464-14192, 400-03-330, 480-04-004, 400-07-080, 911-09-032, 451-06-004, 451-08-026, 451-10-005), the BBRMI-NL -financed BIOS Consortium (NWO 184.021.007), the Netherlands Organization for Health Research and Development (ZonMW 3100.0038, 940-37-024, 31160008), EMGO+ Institute for Health and Care Research, Neuroscience Campus Amsterdam, BBMRI–NL (184.021.007: Biobanking and Biomolecular Resources Research Infrastructure), National Institutes of Health (NIH 5R37DA018673–03, R01 MH059160, 1RC2 MH089951-01, 4R37DA018673-06, 1R01 MH087646-01A1), National Institute of Mental Health (RFA MH08120), Brain and Behavior Research Foundation (2011 NARSAD Distinguished Investigator Grant; 18633), FP7 ENGAGE (FP7-HEALTH-F4-2007-201413), European Research Council (230374-GMI, 284167), Rutgers University (3797).
DTR: This work was supported by the Integrated research on DEvelopmental determinants of Aging and Longevity (IDEAL), an EU's FP7 project with number: 259679. The Danish Twin Registry is supported by a grant from The National Program for Research Infrastructure 2007 from the Danish Agency for Science Technology and Innovation.






