1. Introduction
Transposable elements are important constituents of the genomes of most organisms. When they become active, they can cause deleterious mutations and chromosome breaks. It therefore stands to reason that mechanisms would have evolved to prevent or limit their activity. Recent studies have pointed to the phenomenon of RNA-mediated silencing as one of these mechanisms (Blumenstiel & Hartl, Reference Blumenstiel and Hartl2005; Brennecke et al., Reference Brennecke, Aravin, Stark, Dus, Kellis, Sachidanandam and Hannon2007, Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008; Josse et al., Reference Josse, Teysset, Toideschini, Sidor, Anxolabéhère and Ronsseray2007; Das et al., Reference Das, Bagijn, Goldstein, Woolford, Lehrbach, Sapetschnig, Buhecha, Gilchrist, Howe, Stark, Matthews, Berezikov, Ketting, Tavaré and Miska2008; Jensen et al., Reference Jensen, Stuart, Goodpaster, Goodman and Simmons2008).
In the fruit fly Drosophila melanogaster, RNA-mediated silencing has been implicated in the regulation of different kinds of transposable elements, including the I factor, which is a retrotransposon, and the P element, which is a cut-and-paste transposon (Reiss et al., Reference Reiss, Josse, Anxolabéhère and Ronsseray2004; Dramad et al., Reference Dramad, Heidmann and Jensen2007; Simmons et al., Reference Simmons, Niemi, Ryzek, Lamour, Goodman, Kraszkiewicz and Wolff2007; Brennecke et al., Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008; Chambeyron et al., Reference Chambeyron, Popkova, Payen-Groschêne, Brun, Laouini, Pelisson and Bucheton2008). Both of these transposable elements were discovered through their involvement in a syndrome of abnormal traits in the offspring of crosses between different strains. This syndrome, called hybrid dysgenesis, usually occurs in the offspring of only one of the two reciprocal crosses (Kidwell et al., Reference Kidwell, Kidwell and Sved1977; Bregliano et al., Reference Bregliano, Picard, Bucheton, Pelisson, Lavige and L'Heritier1980; Engels, Reference Engels, Berg and Howe1989). In the offspring of the other cross, dysgenesis is repressed by a maternally transmitted condition now thought to depend on small RNA molecules that interfere with the expression or functioning of transposon RNA.
In this paper, we focus on the repression of P-element-induced hybrid dysgenesis. This type of dysgenesis is restricted to the germ line because the transposase that mobilizes P elements is naturally made only in germ-line cells. P-induced dysgenesis is manifested by the frequent occurrence of mutations and by the failure of the gonads to develop. This latter effect, called gonadal dysgenesis (GD), is a profound abnormality that causes the flies to be sterile. P-induced dysgenesis can be repressed by a maternally inherited condition called the P cytotype (Engels, Reference Engels1979a). A series of studies has tied this repressing condition to P elements that have fortuitously inserted in the telomere associated sequences (TAS) at the left end of the X chromosome (Ronsseray et al., Reference Ronsseray, Lehmann and Anxolabéhère1991, Reference Ronsseray, Lemaitre and Coen1993, Reference Ronsseray, Lehmann, Nouaud and Anxolabéhère1996, Reference Ronsseray, Marin, Lehmann and Anxolabéhère1998; Marin et al., Reference Marin, Lehmann, Nouaud, Izaabel, Anxolabéhère and Ronsseray2000; Stuart et al., Reference Stuart, Haley, Swedzinski, Lockner, Kocian and Simmons2002; Niemi et al., Reference Niemi, Raymond, Patrek and Simmons2004; Simmons et al., Reference Simmons, Raymond, Niemi, Stuart and Merriman2004; Jensen et al., Reference Jensen, Stuart, Goodpaster, Goodman and Simmons2008). This locus is known to produce small RNA molecules that interact with proteins of the PIWI family, and for that reason, its RNA products are called PIWI-interacting, or piRNAs (Brennecke et al., Reference Brennecke, Aravin, Stark, Dus, Kellis, Sachidanandam and Hannon2007). Recent studies have suggested that piRNAs are the physical basis of the P cytotype (Brennecke et al., Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008). However the mechanistic details, including how these RNAs are produced and act, remain to be elucidated.
Several P elements inserted in the TAS at the left end of the X chromosome have been isolated and analysed. These elements vary in size, sequence and insertion site. They also appear to differ in their ability to evoke the P cytotype – that is, in their ability to repress hybrid dysgenesis. For example, the 1·9 kilobase (kb)-long element TP6 is reported to be a stronger repressor of GD than the 1·8 kb-long element TP5. However, in an assay that measured the frequency of P-element excisions from the X-linked singed (sn) locus, TP5 was the stronger repressor (Stuart et al., Reference Stuart, Haley, Swedzinski, Lockner, Kocian and Simmons2002). It is not clear if these differences are due to specific features of the elements or to differences in the genomic DNA around them. Drosophila telomeres are dynamic structures, differing one from another in length and sequence composition (Mason & Biessmann, Reference Mason and Biessmann1995). These differences can be seen within the TAS, where telomeric P elements are inserted, and also in the array of retrotransposons that forms the portion of the telomere distal to the TAS. Such differences might influence the ability of sequences within the TAS to produce piRNAs.
Recent studies have suggested that piRNAs – for the I factor as well as for the P element – are transmitted maternally (Brennecke et al., Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008). These RNAs may therefore explain the cytoplasmic component of transposon regulation that has been documented in some genetic analyses. Specifically, daughters that do not inherit a telomeric P element (TP) from their heterozygous TP/+ mothers repress GD about as effectively as daughters that do inherit such an element (Simmons et al., Reference Simmons, Niemi, Ryzek, Lamour, Goodman, Kraszkiewicz and Wolff2007). Thus repression can be mediated by a strictly maternal effect of the TP. However, no such effect is seen when the dysgenic trait studied is P-element excision in males (Stuart et al., Reference Stuart, Haley, Swedzinski, Lockner, Kocian and Simmons2002).
In this paper, we use stocks that contain either TP5 or TP6 to address several questions about cytotype regulation. One question is whether variation in repression ability is consistent across different genetic assays. Do telomeric P elements that strongly repress increased mutability also strongly repress sterility? Another question is whether variation in repression ability is due to structural differences among the telomeric P elements, or to contextual differences in the telomeric DNA. Does repression depend strictly on the P element's structure and sequence, or does it depend on the overall structure of the telomere into which the P element is inserted? We also ask if the strength of cytotype regulation is related to P-element expression. A telomeric P element might be expected to produce piRNAs at the expense of mRNAs, and these piRNAs might be expected to target transposase mRNA for destruction. Is strong P-element regulation correlated with weak mRNA production from the telomeric P element, and is this regulation associated with a reduction in the amount of transposase mRNA? Finally, we ask what circumstances allow the maternal component of cytotype regulation to contribute to the repression of P elements in the offspring of dysgenic crosses.
2. Materials and methods
(i) Drosophila stocks and husbandry
Information on the special chromosomes and mutant alleles used in the experiments is available on the Flybase website, in Lindsley & Zimm (Reference Lindsley and Zimm1992), or in other references cited in the text. Experimental cultures were reared in vials on a standard cornmeal–molasses–dried yeast medium at 25°C except where noted.
The X-linked telomeric P elements TP5 and TP6 were genetically isolated from all the other P elements in two wild-type strains in late 1997, as described by Stuart et al. (Reference Stuart, Haley, Swedzinski, Lockner, Kocian and Simmons2002). In January 1998, these elements were established in homozygous stocks that also carried the linked markers w, m and f. Secondary stocks were set up from these primary stocks to serve as reserves; thereafter the primary and secondary stocks were maintained independently by mass transfers of adults each generation. In March 1999, homozygous TP5 w snw and TP6 w snw stocks were established by recombining X chromosomes from the primary TP stocks with an X chromosome carrying the double P-element insertion mutation singed-weak (snw), as described by Stuart et al. (Reference Stuart, Haley, Swedzinski, Lockner, Kocian and Simmons2002); we refer to these recombinants as the primary TP snw stocks. In September 2004, following the same procedure, additional TP snw stocks were established by recombining X chromosomes from the secondary TP5 and TP6 stocks with an snw X chromosome; we refer to these later recombinants as the secondary TP w snw stocks and denote them as TP5′ snw and TP6′ snw. Each of the TP snw and TP′ snw stocks was started with a single recombinant X chromosome. In addition to the telomeric P elements and the P elements inserted in the singed gene, these recombinant chromosomes carried a small P element tightly linked to the singed locus. This element, near but not within the singed gene, has been called the ‘unsinged’ P element (Roiha et al., Reference Roiha, Rubin and O'Hare1988).
(ii) GD assay for repression of P-element activity
GD is the most severe manifestation of P-element activity in the germ line. To test for repression of this trait, females from a stock or a particular cross were mated to males from the P strain Harwich (Kidwell et al., Reference Kidwell, Kidwell and Sved1977), which is a strong inducer of dysgenesis, at 21°C. After 3 days, each mated female was transferred to a fresh culture, which was incubated for 11 days at 29°C, a temperature that enhances the occurrence of GD. The progeny were transferred to a holding vial and allowed to mature for 2 days, and then as many as 20 of the females of each genotype were scored for the presence or absence of eggs. The procedure was to squash the females between two glass slides in a solution of diluted food colouring, which helps to visualize the eggs. Females without any eggs were scored as having GD; those with at least one egg were scored as normal (non-dysgenic). Repression of P activity was indicated by a low frequency of GD.
(iii) Mutability assay for repression of P-element activity
The singed-weak (snw) allele causes a mild malformation of the bristles on the adult cuticle. This allele is hypermutable because the two P elements inserted in it are frequently excised by the action of the P transposase (Engels, Reference Engels1979b, Reference Engels, Berg and Howe1989). Excison of one P element produces an allele with a more extreme mutant phenotype (sne) and excision of the other P element produces an allele with a pseudo-wild phenotype (sn(+)). With natural transposase sources, these excisions occur only in the germ line. To detect them, the flies in which the excisions are occurring must be crossed appropriately. Individual snw males were crossed to C(1)DX females that had attached-X chromosomes and their sons, which inherited an X chromosome patroclinously, were scored for the three possible bristle phenotypes. The proportion of sne and sn(+) flies among those scored was used to estimate the frequency of P excisions in the germ lines of the tested males. Individual snw/+ females were crossed to sn3 males and their offspring (both sons and daughters) were scored for the snw and sne phenotypes. Flies that are heterozygous for sn3 and an sne allele derived from snw have the sne phenotype and flies that are heterozygous for sn3 and snw have an snw phenotype. We did not count the sn+ flies in these cultures because of the pre-existing sn + allele in the tested females. The proportion of sne flies among those scored was used to estimate the frequency of P excisions in the female germ line. Obviously the estimates for P excision frequency in males and females are not equivalent. In all the snw mutability experiments, the test cultures were scored on days 14 and 17 after they were established. Repression of P activity was indicated by a low frequency of P excisions.
Three sources of the P transposase were used in the snw mutability tests: (1) the P(ry +, ▵2–3)99B transgene inserted on chromosome III (Robertson et al., Reference Robertson, Preston, Phillis, Johnson-Schlitz, Benz and Engels1988), hereafter denoted as ▵2–3, (2) the H(hsp/CP)2 transgene inserted on chromosome II (Simmons et al., Reference Simmons, Haley, Grimes, Raymond and Niemi2002), hereafter denoted as CP and (3) the Harwich P strain, marked either with w (the standard Harwich strain) or with y and w (a strain that was created by incorporating the y marker into the standard strain through a series of backcrosses). When the ▵2–3 transgene was used, the test males were crossed to attached-X females from a P strain to repress the bristle mosaicism that is caused by ▵2–3's ability to destabilize snw outside the germ line (Robertson & Engels, Reference Robertson and Engels1989).
(iv) Statistical analyses
The experiments to study repression of GD or snw mutability included many replicate cultures of each test group. All the data for an experiment were collected within a 1- or 2-week period. Unweighted mean frequencies of GD and P excisions from snw were calculated by treating all replicate measurements equally, and the variances associated with these means were calculated empirically. Statistical differences between groups within experiments were assessed by t- or z-tests using standard errors of the unweighted sample means.
(v) RNA isolation and reverse transcription (RT)-PCR
RNA was isolated from groups of 20 whole, virgin females using TRIZOL (Invitrogen) according to the supplier's instructions. RNA pellets were rehydrated in 20 μl diethylpyrocarbonate (DEPC)-treated water and two 4 μl portions from each sample were removed for analysis. One portion was subjected to RT by the ThermoScript reverse transcriptase (Invitrogen) using an oligo-dT primer in a total volume of 20 μl according to the supplier's instructions. The other portion was added to 16 μl of DEPC-treated water to serve as a non-RT control. After RT was completed, both RT and control samples were treated with 1 μl RNase A (10 mg/ml) to clear them of RNA. The DNA in 1 or 2 μl portions from these samples was then amplified by the PCR in a total volume of 25 μl using appropriate primers and temperature profiles; see Jensen et al. (Reference Jensen, Stuart, Goodpaster, Goodman and Simmons2008) for details. The PCR products were analysed on 1% agarose gels run at 70 volts.
3. Results
(i) Variation in repression of hybrid dysgenesis
The primary and secondary TP snw stocks were tested for repression of GD and snw mutability shortly after the secondary stocks were established. The P strain Harwich was used to induce GD, and the ▵2–3 transgene was used to induce snw mutability. Three years later, the TP snw stocks were re-tested for repression of snw mutability using the CP transgene as a source of the P transposase. A w snw strain that did not carry a telomeric P element was included as a control in each of these experiments. The results are summarized in Table 1.
Table 1. Repression of GD and snw mutability by primary and secondary TP snw stocks

GD was induced in the daughters of crosses between males from the standard Harwich strain (marked with w) and stock females. snw mutability was induced in the sons of crosses between ▵2–3 or CP males and stock females. With ▵2–3 the initial crosses were at 21°C and with CP they were at 25°C; however, in both cases, the testcrosses to C(1)DX females were carried out at 25°C. The CP snw mutability data were collected in conjunction with studies of TP and CP RNA levels (see Fig. 1). Significant repression of GD or snw mutability, relative to the w snw control, is indicated by an asterisk.
a See ‘Materials and methods’ section for the origin of the primary and secondary TP snw stocks; the secondary stocks are denoted with primes.
b Unweighted average percentage of GD among females±standard error.
c Unweighted average frequency of sn(+) and sne sons among all sons±standard error.
In the initial tests, three of the four TP snw strains repressed GD and snw mutability significantly. The only non-repressor was TP5′ snw. However, in the later test this strain showed a moderate, but significant, ability to repress snw mutability, possibly because the CP transgene produces less transposase than the ▵2–3 transgene (compare the control levels of snw mutability in these two experiments; also see Simmons et al., Reference Simmons, Haley, Grimes, Raymond and Niemi2002). In all three experiments, the rank order of the four strains was the same: TP5 snw (strongest repressor), TP6′ snw, TP6 snw and TP5′ snw (weakest repressor). Therefore, the results of the three repression assays are consistent even though they involve different phenotypes (GD and snw mutability), transposase sources (P strain, ▵2–3 transgene and CP transgene) and sexes (GD in females and snw mutability in males), and were carried out at different times.
One possible explanation for the weaker repression ability of the TP5′ snw and TP6 snw strains is that the telomeric P elements had been lost from some of the X chromosomes in them. To test this possibility, PCR was used to screen for these P elements in 15 males from each strain. Genomic DNA was extracted from each male separately using the procedure of Gloor & Engels (Reference Gloor and Engels1992) and then TP5- and TP6-specific primers were used in conjunction with a primer near the 3′ end of the P element (‘3′-in’) to amplify the TP DNA; see Stuart et al. (Reference Stuart, Haley, Swedzinski, Lockner, Kocian and Simmons2002) for details. A product indicating the presence of the telomeric P element was obtained in all the amplification reactions. Thus, the low repression ability of the TP5′ snw and TP6 snw strains was not due to the loss of the telomeric P element.
(ii) Molecular correlates of strong and weak repression
The TP5 snw and TP5′ snw strains provided an opportunity to examine strong and weak repression at the molecular level. We used RT-PCR to assess the relative amounts of different mRNAs in flies derived from these strains. Total RNA was extracted from females obtained by crossing each of the strains with males homozygous for the CP transgene. These females were sisters of the males that were tested for snw mutability (right side of Table 1). The polyadenylated RNA in each extract was reverse transcribed into cDNA, and then pairs of primers were used to amplify samples of the cDNA via PCR. The products from these amplifications were assayed by gel electrophoresis and ethidium bromide staining. Eight independent extracts were prepared and processed from each of the two different genotypes (TP5 snw/+; CP/+ and TP5′ snw/+; CP). In the gels, a sample from one genotype was placed beside a sample from the other to permit pairwise comparisons. Figure 1 presents the results of these RT-PCR experiments.
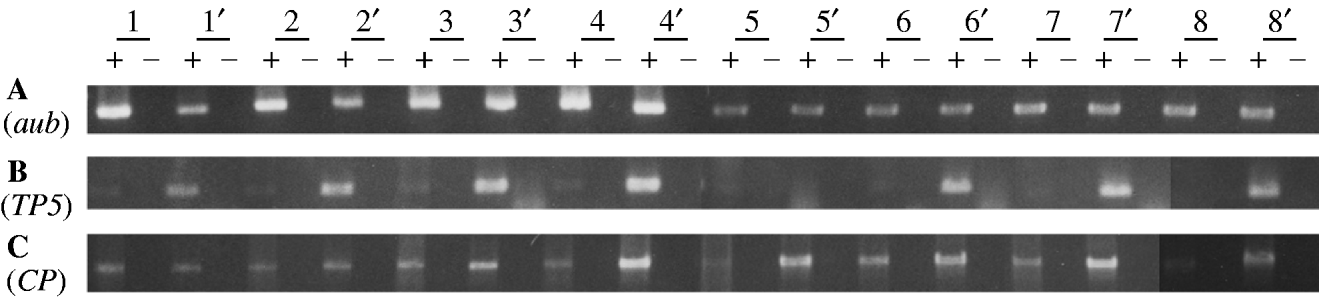
Fig. 1. RT-PCR analysis of germline mRNA from the TP5 snw (strong repressing) and TP5′ snw (weak repressing) strains. Each strain is represented by eight independently obtained samples, with those from the TP5′ snw strain denoted by a prime. Information on the reagents and conditions for the PCR amplifications is given in Jensen et al. (Reference Jensen, Stuart, Goodpaster, Goodman and Simmons2008). A plus denotes where a sample has been reverse transcribed, and a minus denotes where it has not. (A) Amplification over 25 cycles using primers Aub-d and Aub-u to detect an 848-bp product from aubergine mRNA. (B) Amplification over 30 cycles using primers TP5-d and P▵2/3-u to detect a 471-bp product from TP5 germ-line mRNA. (C) Amplification over 30 cycles using primers P0/1-d and P▵2/3-u to detect a 1495-bp product from CP germ-line mRNA.
Panel A in Fig. 1 shows the products obtained with primers specific for aubergine, a gene known to be expressed in the female germ line. Among the eight pairwise comparisons, there is no consistent difference between the TP5 snw and TP5′ snw samples – therefore no consistent difference in aub mRNA abundance in the TP5 snw/+; CP/+ and TP5′ snw/+; CP/+ genotypes. This result implies that overall the TP5 snw/+; CP/+ and TP5′ snw/+; CP/+ extracts contained equivalent amounts of mRNA, and that whatever differences exist among them are due to random variation.
Given the overall equivalence of the two types of samples with respect to total mRNA, we performed experiments to see if the TP5 snw/+; CP/+ and TP5′ snw/+; CP/+ genotypes differed systematically with respect to specific P mRNAs. Panel B shows the products of RT-PCR with primers specific for germ-line mRNA from the TP5 element. The specificity of this amplification arises from the fact that one primer straddles the deletion breakpoints in the TP5 element and the other primer straddles an intron that is removed from P RNA only in germ-line cells. Thus, panel B provides information about the abundance of TP5 RNA exclusively in the germ line, which is the physiologically relevant tissue for studying P-element regulation. Only trace amounts of the PCR product were detected in the samples from the TP5 snw/+; CP/+ flies. Significantly more product was detected in seven of the TP5′ snw/+; CP/+ samples. Under the assumption of equivalent amounts of mRNA in the two types of samples, this result or a more extreme one should occur with probability 0·035. Thus, by a one-sided paired comparison test, we must reject the hypothesis that the two types of samples contain equivalent amounts of TP5 mRNA in the germ line. The consistent difference observed between the TP5 snw/+; CP/+ and TP5′ snw/+; CP/+ RNA samples indicates that strong repression of P-induced hybrid dysgenesis is associated with weak expression of mRNA from the TP5 element.
Jensen et al. (Reference Jensen, Stuart, Goodpaster, Goodman and Simmons2008) provided evidence that cytotype repression of hybrid dysgenesis involves a reduction in the amount of transposase-encoding P mRNA. In the material at hand, this RNA is produced by the CP transgene. Panel C in Fig. 1 shows the products of RT-PCR with primers specific for CP mRNA in the germ line. In this amplification one primer straddles an intron present in CP but not in TP5, and the other primer straddles the P intron that is removed only in germ-line cells. The binding site for this latter primer is not present in either of the P elements inserted in the snw allele, or in the ‘unsigned’ element tightly linked to snw. Thus, DNAs derived from any of these elements are not amplified in reactions with this primer. In seven of the eight pairwise comparisons between samples from the strong and weak repressing genotypes, germ-line CP mRNA was less abundant in the strong repressing sample. This statistically significant result indicates that the strength of repression is mechanistically related to a reduction in the amount of transposase-encoding mRNA.
(iii) Maternal transmission of repression ability
Cytotype regulation by telomeric P elements involves a maternal effect. To investigate this effect, we measured the incidence of dysgenic traits in the offspring of females that were heterozygous for a telomeric P element. These females were created by crossing females from the various TP snw stocks to y snw males; the TP snw stocks all carry the y + allele of the y locus for body colour. Dysgenesis was then induced by crossing the TP snw/y snw F1 females to Harwich y w males. Among the F2 offspring of these crosses, the presence or absence of the telomeric P element was ascertained by following the body colour locus, which is very tightly linked to the left telomere of the X chromosome – and therefore to the TP insertion site; the offspring with grey bodies carried the y + allele and the telomeric P element and those with yellow bodies did not. By observing the level of dysgenesis in the two types of offspring and in controls created by using the w snw stock, we could determine if repression was mediated by a strictly maternal effect of the telomeric P element. Table 2 summarizes the results of these experiments.
Table 2. Repression of P strain-induced hybrid dysgenesis in the offspring of TP snw/y snw females that had homozygous TP snw mothers
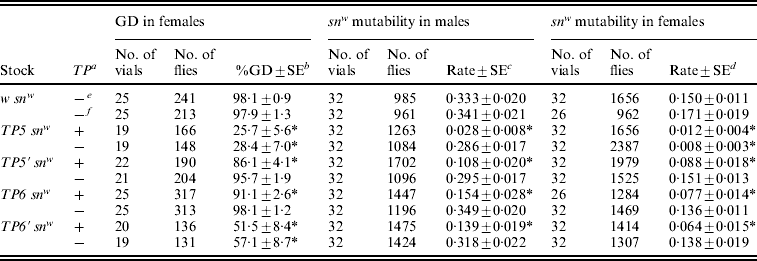
The tested offspring were obtained in a two-generation scheme. Stock females were crossed to y snw males at 25°C and the TP snw/y snw F1 daughters were then crossed to Harwich y w males to induce hybrid dysgenesis. One set of F1 cultures was reared at 29°C and the resulting F2 females were scored for GD; the females with grey bodies carried the TP, whereas those with yellow bodies did not. A second set of F1 cultures was reared at 21°C. F2TP snw (grey) and y snw (yellow) sons from these cultures were individually crossed to C(1)DX, y w f females to produce F3 flies, which were scored to obtain the data on snw mutability in the male germ line. F2TP snw/y sn + (grey) and y snw/y sn + (yellow) daughters from these same F1 cultures were individually crossed to sn3 males to produce F3 flies, which were scored to obtain the data on snw mutability in the female germ line. Significant repression of hybrid dysgenesis relative to the corresponding w snw control is indicated by an asterisk.
a TP present (+) or absent (−) in F2 flies.
b Unweighted average percentage GD±standard error.
c Unweighted average frequency of sn(+) and sne sons among all sons±standard error.
d Unweighted average frequency of sn(e) offspring among snw and sne offspring±standard error.
e F2 flies with grey bodies.
f F2 flies with yellow bodies.
Three dysgenic traits were monitored: GD in females, snw mutability in males and snw mutability in females. All three traits were repressed in flies that inherited a telomeric P element from the heterozygous F1 females, no matter which strain the element came from. This type of repression reflects both maternal and zygotic effects of the telomeric P element. The greatest repression by these combined effects was seen in the flies derived from the TP5 snw strain, which was known to be the strongest repressor (Table 1). Much less effective repression was seen in the flies derived from the other strains. Significant repression by a strictly maternal effect was detected only in tests involving females. GD was repressed in females from both TP5 snw and TP6′ snw (the two strongest repressing strains according to Table 1), and snw mutability was repressed in females from TP5 snw.
More than a year after obtaining the results in Table 2, we used RT-PCR to assess the level of transposase-encoding mRNA in females that might show the maternal effect. In this experiment, RNA was isolated from six replicate samples of each of three female genotypes: (1) TP5 snw/y sn +; CP/+ and (2) y snw/y sn +; CP/+, both derived from a cross between TP5 snw/y snw females and y sn +; CP males and (3) y snw/y sn +; CP/+, which came from a cross between +snw/y snw females and y sn +; CP males. This last genotype served as a control. The polyadenylated RNA in each sample was reverse transcribed into cDNA and then PCR was performed to amplify specific cDNAs. No consistent differences were seen among the three types of samples in amplifications with primers for the aubergine gene (Fig. 2, panel A). However, amplification with primers specific for germ-line CP cDNA revealed that the samples from the TP5 snw/y sn +; CP/+ females consistently had less product than the samples from the control genotype (Fig. 2, panel B). These results indicate that germ-line CP mRNA is diminished by the combined maternal and zygotic effects of the TP5 element – a result that accords with the findings in Fig. 1. There was, however, no consistent evidence for a reduction in germ-line CP mRNA by a strictly maternal effect of the TP5 element. Only two of the six samples from the y snw/y sn +; CP/+ females that had TP5 snw/y snw mothers showed less RT-PCR product than the corresponding control samples.
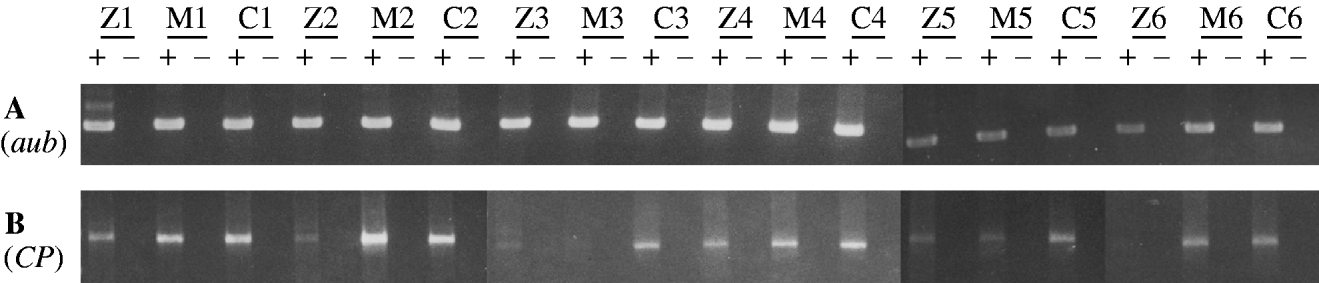
Fig. 2. RT-PCR analysis of germline mRNA from the daughters of crosses between TP5 snw/y snw or +snw/y snw females and y sn +; CP males. RNA was independently obtained from six samples of each of three different genotypes: TP5 snw/y sn +; CP/+, reflecting the zygotic (Z) and maternal effects of the TP5 element from the strong repressing TP5 snw strain; y snw/y sn +; CP/+, reflecting the strictly maternal (M) effects of this element; and y snw/y sn +; CP/+, a control lacking any effect of the TP5 element; see text for details. A plus denotes where a sample has been reverse transcribed, and a minus denotes where it has not. (A) Amplification over 25 cycles using primers Aub-d and Aub-u to detect an 848-bp product from aubergine mRNA. (B) Amplification over 30 cycles using primers P0/1-d and P▵2/3-u to detect a 1495-bp product from CP germ-line mRNA.
In connection with this molecular analysis, we also performed genetic tests for repression of GD in females and snw mutability in males and females (Table 3). All the tests provided evidence for significant repression by a combination of maternal and zygotic effects. However, only the GD tests showed significant repression by a maternal effect alone. The snw mutability tests hinted at such an effect, but the reductions in mutability seen in males and females were not statistically significant.
Table 3. Repression of P strain-induced GD and CP-induced snw mutability in the offspring of TP5 snw/y snw females that had homozygous TP5 snw mothers
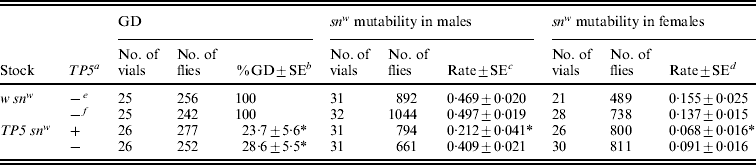
In these experiments, stock females were crossed to y snw males at 25°C and their TP snw/y snw F1 daughters were then either crossed to Harwich y w males at 29°C to induce GD or to y sn +; CP males at 25°C to obtain flies for the snw mutability tests and RT-PCR assays (see Fig. 2). For the snw mutability tests, F2TP snw (grey) and y snw (yellow) males were individually crossed to C(1)DX females and TP snw/y sn + (grey) and y snw/y sn + (yellow) females were individually crossed to sn3 males to produce the F3 flies that were scored. Significant repression of GD or snw mutability relative to the corresponding w snw control is indicated by an asterisk.
a TP present (+) or absent (−) in F2 flies.
b Unweighted average percentage GD±standard error.
c Unweighted average frequency of sn(+) and sne sons among all sons±standard error.
d Unweighted average frequency of sn(e) offspring among snw and sne offspring±standard error.
e F2 flies with grey bodies.
f F2 flies with yellow bodies.
4. Discussion
Extensive data now indicate that the P cytotype is created by P elements that have inserted in the TAS at the left end of the X chromosome. Several different P element insertions in this locus have been identified and analysed.
For example, TP5 and TP6 are inserted in the same orientation at the same nucleotide position within one of the repeat sequences in the TAS; however, these elements differ in size – TP6 is 101 bp longer, and in sequence – TP5 is missing base pairs 438 through 1523, whereas TP6 is missing base pairs 833 through 1816 from the 2907 base pairs of the canonical P element (Stuart et al., Reference Stuart, Haley, Swedzinski, Lockner, Kocian and Simmons2002). Strains homozygous for either of these telomeric P elements vary in their ability to repress hybrid dysgenesis. Among the four strains studied here, one TP5 strain was the strongest repressor and the other TP5 strain was the weakest. The two TP6 strains lay between these extremes. The consistent ranking of these four strains across three different repression assays argues that variation in repression ability cannot simply be due to structural differences between the TP5 and TP6 elements. Rather, it must involve some other factor such as the overall structure of the telomere into which each element is inserted.
Drosophila telomeres consist of two distinct regions: the proximal TAS (Karpen & Spradling, Reference Karpen and Spradling1992), comprising a variable number of repeating units, which are themselves variable in nucleotide sequence, and a distal array of retrotransposons (Mason & Biessmann, Reference Mason and Biessmann1995). The retrotransposon array is capped at its end – the chromosome's end – by a protein complex that includes heterochromatin protein 1 (HP1), the product of the Su(var)205 gene (Fanti et al., Reference Fanti, Giovinazzo, Berloco and Pimpinelli1998). Over time, the retrotransposon array changes. Sequences are lost because of incomplete DNA replication at the end of the chromosome, and sequences are gained when retrotransposons are added to the array. Because of these changes – and possibly others within the TAS, the telomeres in independently maintained stocks are expected to differ from one another, and such differences are known to have functional correlates. Thus, for example, extension of the retrotransposon array can enhance the expression of a transgene inserted within the TAS (Golubovsky et al., Reference Golubovsky, Konev, Walter, Biessmann and Mason2001). Evolving telomere structure may therefore explain why particular TP5 and TP6 strains differ in their ability to repress hybrid dysgenesis – for example, why Stuart et al. (Reference Stuart, Haley, Swedzinski, Lockner, Kocian and Simmons2002) found that in a pair of TP5 snw and TP6 snw strains, the TP5 snw strain was the better repressor of snw mutability, whereas in a pair of TP5 sn + and TP6 sn + strains, the TP6 sn + strain was the better repressor of GD.
To investigate the molecular correlates of these differences, we used RT-PCR to assess the relative abundance of germ-line P mRNAs in females derived from the strong and weak repressing TP5 strains. Females from the weak repressing strain produced more TP5 mRNA in the germ line than their counterparts from the strong repressing strain. The mechanism of repression therefore cannot involve a polypeptide encoded by the TP5 mRNA. Jensen et al. (Reference Jensen, Stuart, Goodpaster, Goodman and Simmons2008) reached this same conclusion on the basis of a different argument. Instead of repressing through polypeptide products, telomeric P elements are now thought to repress hybrid dysgenesis by generating non-coding piRNAs that are primarily antisense in orientation (Brennecke et al., Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008). These molecules are presumably synthesized from antisense transcripts that originate from a promoter within the TAS downstream of the inserted P element. This downstream antisense promoter might have been impaired in the weak repressing TP5′ strain, or the promoter of the TP5 element in this strain might have been upregulated so that sense transcripts are formed in place of antisense transcripts. Such upregulation might be due to the influence of enhancers located in the distal retrotransposon array. Another possibility is that the retrotransposon array in the weak repressing strain initiates transcripts that read sense-ward through the TP5 element. How well a particular telomeric P element represses hybrid dysgenesis may therefore depend on the predominant direction of transcription through that element – determining, ultimately, how much antisense piRNA it can make.
We also examined the levels of transposase-encoding P mRNA in females derived from the strong and weak repressing TP5 strains. The females from the strong repressing strain had less transposase mRNA than the females from the weak repressing strain. This finding suggests that the strength of repression depends on the ability of the piRNAs generated from a telomeric P element to destroy transposase mRNA. Strong repression evidently involves a more vigorous attack by these piRNAs than weak repression.
The Drosophila genome contains many loci that produce piRNAs (Brennecke et al., Reference Brennecke, Aravin, Stark, Dus, Kellis, Sachidanandam and Hannon2007). So far, only one of these loci – in the TAS at the left end of the X chromosome – has been implicated in P-element regulation. A P insertion into this locus allows it to synthesize piRNAs with P-element specificity. In essence, the insertion ‘marks’ the piRNA-producing system for study. Genetic analyses of P regulation by these types of insertions may therefore provide information about the general features of this system. Thus, for example, the ability of a telomeric P element to regulate the P family is established in the female germ-line (Niemi et al., Reference Niemi, Raymond, Patrek and Simmons2004), and once established, this ability can be transmitted by females to their offspring independently of the telomeric P element itself. Patroclinously transmitted telomeric P elements utterly lose their regulatory ability; however, a telomeric P element inherited from a male can reacquire regulatory ability after passing through a female germ line, and this reacquisition is enhanced by the maternal and zygotic effects of another telomeric P element – a phenomenon called presetting (Niemi et al., Reference Niemi, Raymond, Patrek and Simmons2004), or the pre-P cytotype (Ronsseray et al., Reference Ronsseray, Lemaitre and Coen1993).
In terms of the piRNA model of P-element regulation, these genetic findings imply (1) that piRNAs are generated in the female germ line, (2) that they are transmitted to the next generation through the egg cytoplasm and (3) that they play a key role in defending against the P elements that are transmitted paternally in dysgenic crosses. In regard to this last point, Brennecke et al. (Reference Brennecke, Aravin, Stark, Dus, Kellis, Sachidanandam and Hannon2007, Reference Brennecke, Malone, Aravin, Sachidanandam, Stark and Hannon2008) have proposed that antisense piRNAs from a mother's telomeric P element interact with sense mRNAs encoded by paternally transmitted P elements to create a class of sense piRNAs, which then feed back to stimulate the synthesis of more antisense piRNAs, thereby amplifying the regulatory system that represses hybrid dysgenesis. This ‘ping-pong’ mechanism requires that both sense and antisense piRNA sources be present in the genome of the offspring. When the offspring do not inherit the mother's telomeric P, the maternally transmitted piRNAs can provide only a limited defence against dysgenesis. However, if some of the paternally transmitted P elements are able to generate antisense piRNAs – in effect, replacing the telomeric P element that was not inherited maternally, this limited defence can be augmented by ping-pong cycling, starting with maternally inherited antisense piRNAs. This strengthening of P regulation is possible when the paternally transmitted P elements come from a P strain such as Harwich, which, because it has the P cytotype, must contain some P elements that are able to generate antisense piRNAs. However, when the only paternally inherited P element is in the CP transgene, it is difficult to see how more antisense piRNAs could be produced. The absence of a new source of antisense piRNAs may explain why the repression of snw mutability in the y snw/sn +; CP/+ daughters of TP5 snw/y snw mothers and y sn +; CP fathers is modest and not statistically significant, and it may also explain our inability to detect a consistent reduction in the amount of transposase-encoding P mRNA in these daughters.
Males that inherit piRNAs but not a telomeric P element from their mothers do not repress dysgenesis, as assayed by snw mutability, even when they inherit a host of P elements from Harwich fathers. The numerous cell divisions that occur in the germ lines of these males might dilute the maternally inherited piRNAs down to a point at which they are ineffective in preventing P excisions from the snw allele. In addition, the ping-pong mechanism is apparently stymied in these males, possibly because they lack a locus capable of generating antisense piRNAs. In the Harwich genome, the antisense piRNA locus may reside on the X chromosome, which these males did not inherit. By contrast, males that inherit a telomeric P element maternally can become strong repressors of snw mutability. The telomeric piRNA locus therefore is able to function in males. However, this function clearly depends on the locus being inherited maternally because patroclinously transmitted telomeric P elements do not repress snw mutability at all. Perhaps the female germ line imprints the locus in some way, or perhaps maternally transmitted piRNAs are needed to allow its expression in the next generation. In connection with this latter hypothesis, it is known that in females a paternally inherited telomeric P element reacquires its repression ability more fully if the mothers of these females carried a telomeric P element, even when the females do not inherit the element itself (Niemi et al., Reference Niemi, Raymond, Patrek and Simmons2004; Josse et al., Reference Josse, Teysset, Toideschini, Sidor, Anxolabéhère and Ronsseray2007). Maternally inherited piRNAs might therefore be involved in triggering, stabilizing, or enhancing the expression of the telomeric piRNA locus, possibly through an epigenetic alteration of its chromatin.
Jared T. Buschette and Michael P. Goodpaster helped with some of the experiments. The work was supported by funds from the Department of Genetics, Cell Biology and Development and the College of Biological Sciences. M. W. T. received a grant from the Undergraduate Research Opportunities Program of the University of Minnesota.







