The global spread of carbapenemase-producing organisms (CPOs) is an urgent public health threat due to lack effective drugs. Reference Bonomo, Burd and Conly1,Reference Queenan and Bush2 Carbapenemase-producing bacterial pathogens are difficult to treat and are associated with increased severity of illness and mortality. Reference Van Duin and Doi3 The genes encoding carbapenemases are mostly located on mobile genetic elements that could be transferred horizontally to other gram-negative bacteria. Reference Queenan and Bush2,Reference Martínez-Martínez and González-López4 In the United States, Klebsiella pneumoniae carbapenemase (KPC) is the most commonly identified carbapenemase among the carbapenem-resistant Enterobacterales (CRE). Reference Van Duin and Doi3 Oxacillinases (OXA) and other metallo-β-lactamases, such as New Delhi metallo-β-lactamase (NDM), verona integron-encoded metallo-β-lactamase (VIM), and imipenamase-producing metallo-β-lactamase (IMP), are not common and are particularly limited to healthcare-associated common-source outbreaks. Reference Van Duin and Doi3,5 NDM and OXA-48 are widespread in southeast Asia, Europe, China, and Africa. Reference Van Duin and Doi3,Reference Avolio, Vignaroli, Crapis and Camporese6,Reference Doi, O’Hara and Lando8 However, carbapenemase-producing carbapenem-resistant Pseudomonas aeruginosa (CP-CRPA) strains are reported rarely in North America, in part because medical laboratories do not routinely test for them.
Although CPOs usually carry single carbapenemase genes, few studies have reported dual or multicarbapenemase coproducing organisms. Reference Han, Shi and Wu7 In the United States, the first case of K. pneumoniae coproducing NDM-1 and OXA-232 was reported in Pittsburgh. Reference Cai, Lee and Kwa9 However, P. aeruginosa and A. baumannii coproducing different carbapenemases has never been reported. Here, we describe an unusual occurrence of patients infected with K. pneumoniae, P. aeruginosa, and carbapenem-resistant A. baumannii (CRAB) coproducing multiple carbapenemases.
Methods
In Texas, clinical isolates identified to be resistant to multiple antibiotics at healthcare facilities are routinely referred to the Houston Health Department (HHD) and Antibiotic Resistant Laboratory network (ARLN) for further analysis. We examined these data for infections caused by different carbapenemase coproducing pathogens from 2018 to 2021. This research followed the first identification of a cluster of KPC-producing CRPA in the greater Houston area and their unusual antimicrobial susceptibility profiles. Bacterial isolates were identified by matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy (MALDI-TOF MS). Confirmation of carbapenemase production was done using the modified carbapenem inactivation method (mCIM). Polymerase chain reaction (PCR) and whole-genome sequencing were used to identify carbapenemase genes: blaKPC, blaNDM, blaOXA-48-like, blaIMP, and blaVIM. Antibiotic susceptibility tests were performed using the broth microdilution method. Isolates showing rare resistance mechanisms were also sent to the Centers for Disease Control and Prevention (CDC) for further analysis. Patient demographics, microbiological, and clinical information were abstracted from the surveillance data and medical records. This study was reviewed and approved by the Institutional Review Board of University of Texas Health Science Center at Houston.
Results
From January 2018 to July 2021, 7 patients infected with multidrug-resistant organisms coproducing multiple carbapenemases were identified. Table 1 shows the demographic, clinical, and other characteristics of the patients. Moreover, 4 patients were infected with CRE K. pneumoniae, 4 were identified with CRPA, and 1 was infected with CRAB. All patients were reported from different healthcare facilities and 5 patients had invasive renal devices (Table 1B).
Table 1. Clinical Risk Factors and Presumed Sources of Infection for the Carbapenemase-Coproducing Organisms

Note. MDRO, multidrug-resistant organism; CKD, chronic kidney disease; HD, hemodialysis; ESRD, end-stage renal disease; CVD, cardiovascular disease; mCIM+, carbapenemase positive by modified carbapenem inactivation method.
a Both case 1 and 2 had an invasive medical procedure abroad in the past month prior to the specimen collection.
Patient 1 had traveled to India and was hospitalized there for renal disease. The treatment in India was unsuccessful, and this patient was admitted to an acute-care hospital in Houston 4 days after arrival in the United States. A urine culture showed NDM and OXA-48 coproducing K. pneumoniae and NDM and IMP coproducing P. aeruginosa. The isolates were resistant to multiple antibiotics, including colistin and polymyxin (Table 2).
Table 2. Antimicrobial Susceptibility Profiles of Carbapenemase Coproducing Organisms Isolated from the Patients
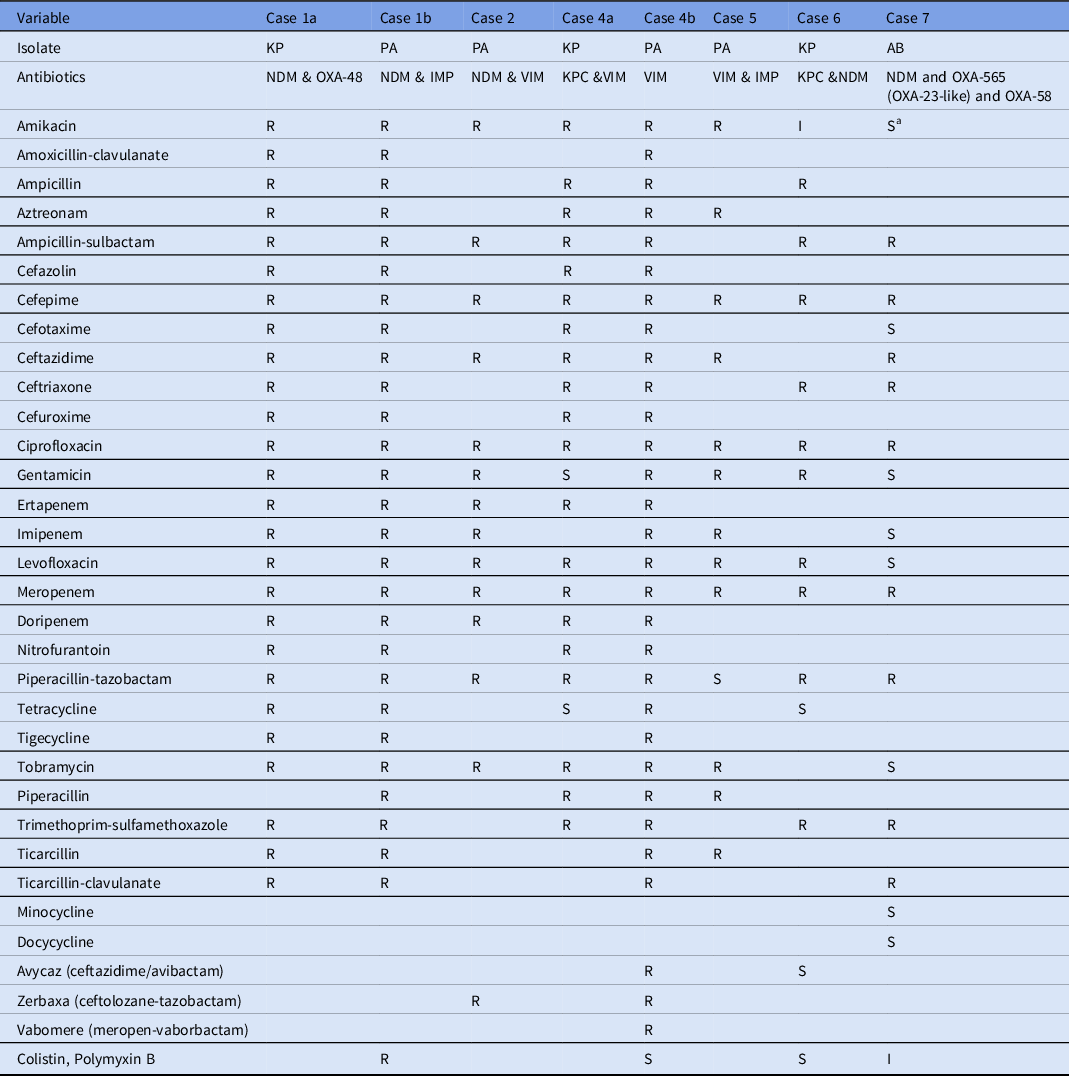
Note. KP, K. pneumoniae; PA, P. aeruginosa; AB, A. baumannii; R, resistant; I, intermediate, S, susceptible.
a Susceptibility report at clinical laboratories showed resistance to Amikacin. For the case 3, complete antibiotic susceptibility profile was not recorded in the surveillance and hence not included in this table.
Patient 2 had a nephrostomy tube for ureteral stricture and hydronephrosis in Nigeria and was admitted to an acute-care hospital 1 week after arrival in the United States. Initial urine culture identified a carbapenemase-positive P. aeruginosa. Whole-genome sequencing, mCIM, and real-time PCR confirmed the presence of VIM and NDM carbapenemases. Antibiotic susceptibility test showed resistance to carbapenems, amikacin, tobramycin, gentamycin, and zerbaxa, a ceftolozane-tazobactam combination.
Patient 3 was admitted to the hospital for a cardiac problem. Urine culture identified K. pneumoniae with KPC and OXA-48 genes. Details of this patient’s medical history and healthcare abroad were not available.
Patient 4 had no history of travel abroad and was initially admitted to an intensive care unit. Wound culture revealed carbapenemase-producing K. pneumoniae harboring both KPC and VIM genes. In addition, a CRPA isolate harboring the VIM gene was also identified. The CRPA-VIM isolate was resistant to all antibiotics tested at the hospital. The patient was readmitted to an acute-care hospital 2 months later, and another urine culture also grew a highly resistant CRPA coproducing a VIM carbapenemase.
Patient 5 was a resident of a rehabilitation facility. CRPA was initially identified from routine urine culture, which was confirmed coproducing VIM and IMP carbapenemases. The patient’s medical history and healthcare abroad was not available.
Patient 6 had liver transplant and end-stage renal disease with a trialysis catheter port but developed intrabdominal complications. The patient had sepsis with K. pneumoniae bacteremia, which was identified to be coproducing NDM and KPC carbapenemases. Furthermore, a blaNDM-1 producing E. coli was also identified.
Patient 7 received outpatient foot wound care and culture from the wound grew carbapenem-resistant A. baumannii. Whole-genome sequencing and mCIM showed the presence of 3 carbapenemases: NDM, OXA-565 (OXA-23 like), and OXA-58. The patient had no recent history of medical care abroad, healthcare facility admission, or invasive medical device.
Discussion
We describe a rare occurrence of multidrug-resistant organisms coproducing multiple carbapenemases in the United States, posing an urgent public health threat. These were K. pneumoniae coproducing KPC and OXA-48, NDM and OXA-48, KPC and VIM, KPC and NDM; P. aeruginosa coproducing NDM and IMP, NDM and VIM, IMP and VIM; and A. baumannii coproducing NDM and OXA variants OXA-565 (OXA-23-like) and OXA-58. Patient 1 was coinfected with K. pneumoniae coproducing NDM and OXA-48, and P. aeruginosa co-producing NDM and IMP carbapenemases. Although carbapenemase-producing P. aeruginosa infections are generally rare in the United States, the occurrence of 3 different carbapenemases in a single patient is unusual. Moreover, OXA-48 and NDM carbapenemases are also considered rare in the United States and their emergence here is alarming.
Two patients received invasive medical care in India and Nigeria, which may suggest potential importation of the infection. Although, the patients had 1 or more comorbidities, most of the cases had invasive medical procedures or devices, organ transplant, and surgery or wound care.
The P. aeruginosa and K. pneumoniae isolates identified to be coproducing different carbapenemases exhibited high resistance to aminoglycosides, including amikacin, tobramycin, and gentamicin, in addition to carbapenems. These findings are in line with previous reports of K. pneumoniae coproducing NDM-1 and OXA-232 and exhibited high level of resistance to amikacin and gentamycin. Reference Cai, Lee and Kwa9
The P. aeruginosa isolate from patient 4 identified with blaVIM was resistant to multiple combinatorial drug therapies, including vabomere (meropenem and vaborbactam), avycaz (ceftazidime and avibactam), and zerbaxa (ceftolozane and tazobactam). These therapies are novel treatment strategies for complicated gram-negative infections. Moreover, the P. aeruginosa isolate from patient 1 coproducing IMP and NDM carbapenemases also exhibited resistance to all antibiotics tested, including colistin and polymyxin B, which are considered last-line drugs for extensively drug-resistant gram-negative infections. Reference Cai, Lee and Kwa9 Although CRAB expressing OXA-23 like and OXA-40 has been previously reported in the United States, Reference Koirala, Tyagi and Guntupalli10 coproduction of multiple variants of OXA-565 (OXA-23-like) and OXA-58 with NDM has never been reported.
Notably, most hospitals and clinical laboratories might not be able to send isolates to public health laboratories for further analysis. Also, not all hospitals may have reported all cases to the health department. Therefore, our data may represent the tip of iceberg.
Acknowledgments
We thank the Houston Health Department, the Antimicrobial Resistance and Laboratory Network, and other referral laboratory centers for testing and molecular characterization of isolates. The findings and conclusions in this study are those of the authors and do not necessarily represent the official position of the Houston Health Department or institutions where the authors are affiliated.
Financial support
This work was supported in part by the National Institutes of Health (NIH R01 grant nos. R01AI116914 and R01AI150685).
Conflict of interest
All authors report no conflicts of interest relevant to this article.




