INTRODUCTION
Dengue fever/dengue haemorrhagic fever (DF/DHF) is a mosquito-borne disease caused by dengue virus (DENV). There are four known serotypes: dengue virus types 1, 2, 3 and 4 (DENV-1 to DENV-4) [Reference Heinz and van Regenmortel1, Reference Thomas, Strickman, Vaughn, Chambers and Monath2]. DF/DHF occurs mainly in tropical or subtropical countries, e.g. in Southeast Asia, the Americas and the Caribbean. In the 1970s, there were a series of DF outbreaks in some of the southern provinces in China. The first outbreak was caused by DENV-4, the second by DENV-1, and the third by DENV-3 [Reference Qiu3]. DF/DHF has been an infectious disease of particular concern in Guangdong province, China, since 1978 when a DF outbreak occurred in Foshan city [Reference Wen, Liang and Du4]. The first DHF case was reported in 1985–1986, in Hainan Island, China [Reference Qiu5, Reference Yang6]. Before this epidemic in China, DF/DHF outbreaks had been reported in Thailand, Vietnam, Indonesia [Reference Kho7, Reference van Peenen8], and Myanmar [Reference Gubler and Monath9]. In the 1990s, large outbreaks of DF occurred in China, with more than 6000 cases reported [10]. In recent years, highly localized and relatively sporadic yearly outbreaks have been observed in China. The DF/DHF outbreaks in Guangdong province were thought to be related to other worldwide outbreaks in that they were caused mainly by imported cases from other countries [Reference Luo11–Reference Ren13]. Previously there had been no evidence of endemic dengue in Guangdong province; however, a report on the DF epidemic in Guangzhou in 2005 suggested that DENV had recently become endemic in some areas in China [Reference Zhang14]. Phylogenetic analysis of the entire sequence of the E gene of DENV-1 is a powerful molecular epidemiological method for the confirmation of endemic and imported cases. Studies of DENV molecular evolution and sequence data were performed to determine the phylogenetic relationships among viruses within each serotype. A threshold of 6% divergence is currently used to separate different genotypes within a dengue serotype [Reference Rico-Hesse15–Reference Chungue17]. Several phylogenetic studies analysing a large number of DENV-1 virus genomic sequences have been published [Reference Zheng12, Reference Rico-Hesse15]. These studies analysed either a 240-nt sequence in the E/NS1 junction region of the genome or a 180-nt sequence in the E gene and suggested that there were potentially five or three DENV-1 virus genotypes, respectively. A recent study, based on nucleotide sequences of the entire E gene, showed that DENV-1 viruses can be classified into five genotypes [Reference Goncalvez18, Reference Laille19]. One study on full-length sequences of DENV-1 showed that DENV-1 viruses can be classified into four genotypes [Reference Nukui20]. In this study, the entire E gene sequence was used to trace the origin of the DENV-1 virus/strain known to have caused the DF outbreak in Guangdong province in 2006.
In the second half of 2006, DF outbreaks occurred in Guangzhou, Yangjiang, Nanhai and Shantou City in Guangdong province, with the most severe epidemic occurring in Guangzhou. The cause of the outbreak was confirmed as DENV-1. A total of 1010 DF cases were reported in Guangdong province (CDC of Guandong Province, unpublished data). As the origin of these outbreaks was not determined by epidemiological investigation, the question arose again as to whether the DF/DHF outbreaks had become endemic in Guangdong province. To answer this question, we isolated DENV from serum samples obtained from patients in these cities that were found to be positive by TaqMan real-time PCR [Reference Ke21]. The E gene of the Guangdong isolates were sequenced and compared to those of DENV strains isolated in recent years in other parts of China and Southeast Asia.
METHODS
Serum samples, cell lines and viruses
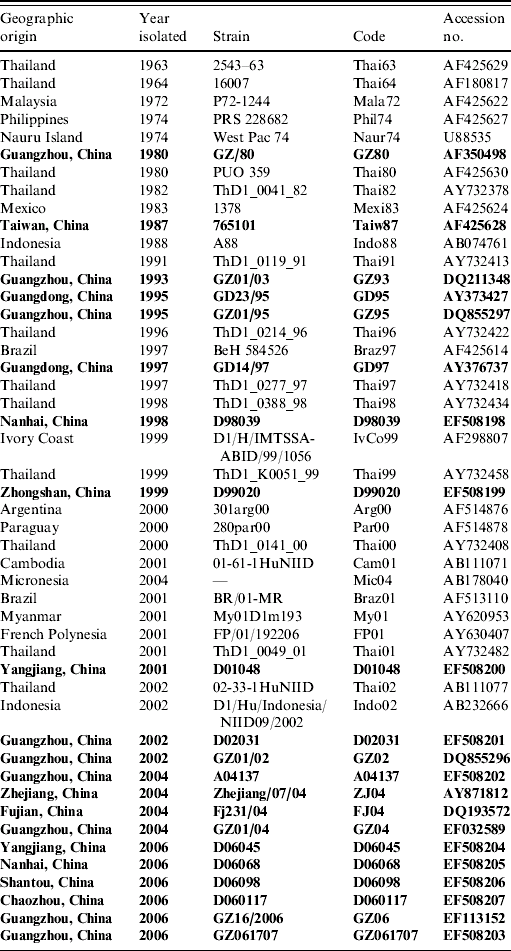
After obtaining informed consent, a total of 103 serum samples were collected from febrile patients during the DF outbreak in Guangdong province in 2006. The mosquito cell line, C6/36, was passaged in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and used for virus isolation. Twenty-three serum samples in which DENV RNAs were found to be positive by real-time PCR [Reference Rico-Hesse15], were inoculated to C6/36 mosquito cells, and 16 isolates were identified as DENV-1. The DENV strains isolated from patients in recent years and which were identified as DENV-1 were also inoculated to C6/36 cells for passaging. The viruses analysed in the present study are listed in Table 1 together with their GenBank accession number and geographic origin.
Table 1. Dengue fever virus strains used in this study
RNA extraction, primer design and reverse transcription–polymerase chain reaction (RT–PCR)
Viral RNAs were extracted from the supernatant of cell culture, to which serum from DF patients or previous DENV isolates had been inoculated and shown a CPE (cell pathogenic effect), using a QIAamp Viral RNA mini kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. To amplify the entire sequence of the DENV E gene, we designed three pairs of primers for RT–PCR with Primer Express version 3.0 (Applied Biosystems, Foster City, CA, USA), as shown in Table 2. RNA samples (5 μl) were used for one-step RT–PCR (TaKaRa, Shiga, Japan). Briefly, the RT reaction was performed at 50°C for 30 min and the RT enzyme was denatured at 95°C for 15 min. Next, the PCR reaction was performed for three cycles at 94°C for 1 min, 60°C for 40 s, and 72°C for 1 min. Subsequently, 30 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 40 s and a final 72°C for 10 min were performed. PCR products were purified with a QIAquick PCR Purification kit (Qiagen) according to the manufacturer's protocol.
Table 2. Primers for amplifying the E gene of DENV type 1

* Based on the numbering of 01-61-1HuNIID strain (accession number AB111071).
Sequencing and genetic analysis
The PCR product was sequenced in both directions using a BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems). Sequencing reaction products were purified with Autoseq G-50 (GE Healthcare–Amersham, Piscataway, NJ, USA), and the nucleotide sequences were determined by ABI Prism® 3100 genetic analyser. The sequences of D1E1, D1E2 and D1E3 were edited using Sequencher™ 4.7 (Gene Codes Corporation, Ann Arbor, MI, USA). The sequences of the DENV-1 E gene amplified in the study were BLASTed against GenBank sequences. The sequences were analysed with the edited sequences of the E gene using Clustal X v.1.83 software. The sequence homologies of nucleic acids and amino acids were calculated and a phylogenetic tree was constructed by the neighbour-joining method using MEGA 3.1 software. One thousand bootstrap repetitions were used for confirmation of the statistical significance of the phylogenetic analysis. The geographic regions from which serum samples were obtained are shown in Figure 1.
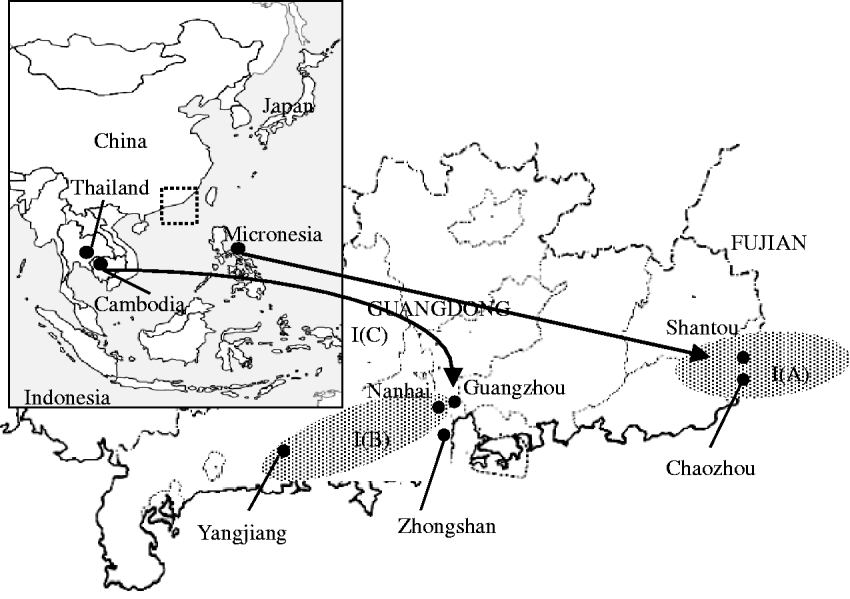
Fig. 1. Geographic representation of locations from which the dengue viruses were isolated. The inset map on the left shows the Southeast Asian region from which the dengue viruses, whose sequences were used in this study, were isolated. The main map shows an enlargement of Guangdong province and the marked areas indicate the locations from which DENV strains were collected. Our hypothesis regarding the origins of the dengue outbreaks is shown schematically. The isolates D06045 and D06068 in cluster B might have originated from a virus imported from Thailand in 2001 before circulating within Guangdong province, while the isolates D06098 and D060117 in cluster A might be derived from a virus imported from Micronesia before circulating within Guangdong province. The two isolates, GZ06 and GZ061707, identified in Guangzhou in 2006, might have two different origins. The former might have come from Thailand and the latter might have originated from Cambodia in 2001. These viruses re-emerged in Guangdong province as endemic in 2006.
RESULTS
Five of 16 DENV-1 isolates obtained from patient serum samples collected during the DF outbreak in Guangdong province in 2006 were sequenced. The sequence of the E gene was determined and analysed phylogenetically along with 43 previously reported isolates (Fig. 2). DENV-1 is divided into five genotypes. The DENV-1 endemic in Southeast Asia consists of genotypes I and IV, and the isolates from Guangdong in 2006 belong to genotype I. For genotype I, seven independent sub-genotypes, A–G, were identified by sequence analysis of the E gene and the homologies of the nucleic acids and amino acids within each sub-genotype were >99%. Sub-genotype A consisted of two isolates from the Guangdong DF outbreak in 2006, Shantou (D06098) and Chaozhou (D060117), and one isolate from Guangzhou (A04137) and one from Fujian (DQ193572) in 2004. An isolate from Micronesia in 2004 (AB178040) also belonged to this sub-genotype. Sub-genotype B contained three DENV-1 isolates from the 2006 DF outbreak in Yangjiang (D06045), Nanhai (D06068) and Guangzhou (GZ06) cities, Guangdong province, and one DENV-1 isolate from 2001 in Yangjiang (D001048) (Fig. 2). Yangjiang and Nanhai cities are located to the west of Guangzhou city and are relatively close to each other (Fig. 1). Interestingly, sub-genotype A also contained a virus isolate AY732482 from Thailand in 2001. Sub-genotype C contained four DENV-1 isolates from Thailand obtained during 1996 or 2000 (Thai96, Thai98 Thai99 Thai00) and one from Nanhai in 1998 (D98039), one from Cambodia in 2001 (Cam01) and one from Guangzhou in 2006 (GZ061707). In sub-genotypes D–G, the isolates were obtained from China and other Southeast Asian countries prior to 1990.
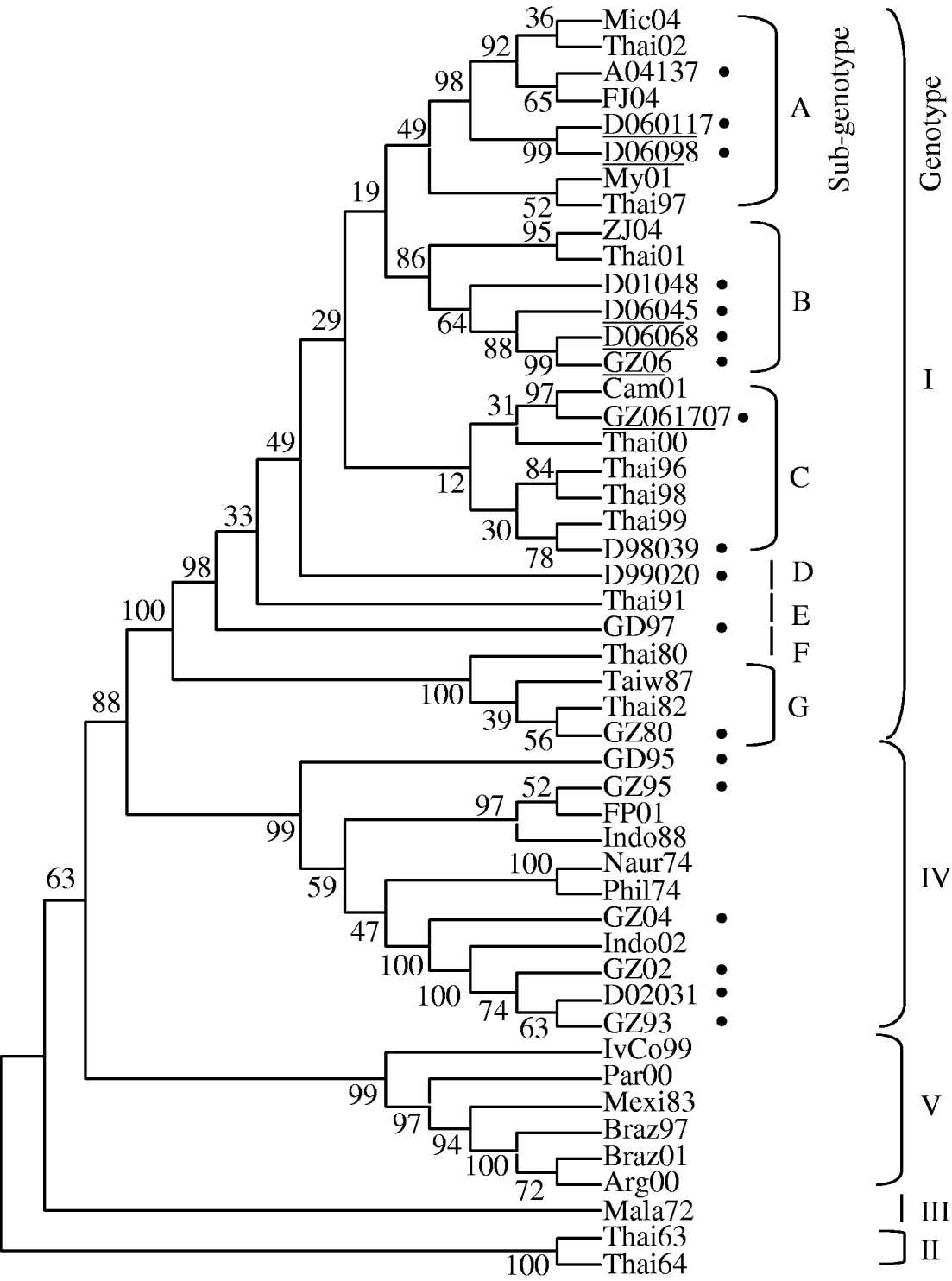
Fig. 2. Phylogenetic analysis of DENV strains isolated during outbreaks, including that in Guangdong province in 2006. The phylogenetic tree is based on the E gene sequences of five isolated DENV strains we sequenced, and 43 sequences from Genbank. The strains isolated in Guangdong province are indicated by black dots (●) to the right of the strain name. The Guangdong isolates are distributed across two genotypes, genotype I and genotype IV, while those in the main sub-genotypes A–G are clustered in genotype I.
DISCUSSION
In previous DF/DHF outbreaks in Guangdong, China, the circulating viruses could be traced back to other countries through the analysis of DENV gene sequences [Reference Zheng12]. Thus, it is believed that the DF/DHF outbreaks in Guangdong were caused mainly by cases imported from Southeast Asian countries around the time of the epidemics in Guangdong [Reference Qiu3–10, Reference Zhang14]. For example, the DF outbreak in Guangzhou, 2002, may have been caused by a case entering the province from Indonesia (Fig. 2, genotype IV), as a DF outbreak was reported at the same time in Indonesia, and the E gene sequences of the viruses isolated from Guangzhou and Indonesia were very similar. Therefore, we hypothesize that the DF epidemics in Yangjiang in 2001 (Fig. 2, sub-genotype B), Nanhai in 1998 (Fig. 2, sub-genotype C), Zhongshan in 1999 (Fig. 2, sub-genotype D), and Guangzhou in 2004 (Fig. 2, sub-genotype A) may have been caused by cases originating in Thailand (for sub-genotype A, and B and C isolates) or Micronesia (Fig. 1).
The scenario for the DF outbreak in Guangdong province in 2006 was different from those of previous outbreaks in Guangdong as the sequence comparison of the E gene of the viruses suggested that the 2006 outbreak was a case of endemic infection of dengue circulating locally in the province in 2006. The E gene sequences of the viruses isolated in Shantou (D06098) and Chaozhou (D060117) were very similar to those isolated in Guangzhou (A04137) and Fujian in 2004 (FJ04) (Fig. 2, sub-genotype A). Although the possibility that the isolated virus may have been imported from another province in 2006 still remains, our hypothesis is that the viruses circulating in Micronesia in 2004 were imported into Guangdong and Fujian provinces in 2004, and re-emerged endemically in Shantou and Chaozhou cities, Guangdong province, in 2006. The E gene sequences of the viruses isolated in Yangjiang and Nanhai cities (D06045 and D06068, respectively) were similar to that of the virus isolated in Yangjiang (D01048) in 2001 (Fig. 2, sub-genotype B). The virus isolated in Yangjiang (D01048) may have been imported from Thailand during a DF epidemic that occurred there in 2001. One of the isolates from Guangzhou in 2006 (GZ061707), together with isolate AB111071 obtained in Cambodia in 2001, belonged to sub-genotype C. No viruses isolated in Guangdong province have been reported with an E gene sequence similar to that isolated in Cambodia in 2001. However, the epidemic might have been caused by an imported virus that had been circulating in Cambodia in 2001 (Fig. 1).
According to our experimental data, we demonstrated the existence of endemic DF cases in the 2006 outbreak in Guangdong province, and that virus strains imported from Southeast Asian countries were also in circulation at the same time.
ACKNOWLEDGEMENTS
This work was supported by Grant-in-aids for Scientific Research and the Programme of Excellence for Zoonosis Control, the 21st Century COE Program, Japanese Ministry of Education, Culture, Sports, Science and Technology (18580301 and 17255009) and Japanese Ministry of Health, Labour and Welfare (H15-Shinkou-17 and H17-Shinkou-ippan-018). We thank Dr H.-M. Wang for his careful revision of the manuscript.
DECLARATION OF INTEREST
None.






