The 2006 Nutrition Prevention of Cancer Trial(Reference Clark, Combs and Turnbull1) reinvigorated interest in assessing human selenium (Se) status and determining human Se requirements because it suggested that Se supplementation would help prevent several forms of cancer. This interest was further enhanced world-wide by concern that UK and European Se intakes were decreasing due to a declining emphasis on imported wheat from North America(Reference Broadley, White and Bryson2). Contemporary reports suggest that UK Se intakes have declined to 29 to 39 μg/d(Reference Rayman3–Reference Rayman5), whereas the current UK Recommended Nutrient Intakes (RNI) are 60 μg and 75 μg/d for adult women and men, respectively(6). In contrast, US median Se intakes were reported to be 98 and 154 μg/d for female and male young adults, respectively, whereas the US dietary reference intake RDAs were modified in 2000 to 55 μg Se/d for both adult women and men(7). In 2005, the New Zealand nutrient reference values for adult women and men were set at 60 and 75 μg/d for adult women and men, respectively(8). These recommendations were all established using differing interpretations of research on the levels of dietary Se necessary to maintain plasma glutathione peroxidase (Gpx3) at plateau levels. In contrast, the WHO recommendation remains at 30 and 40 μg Se/d for women and men, respectively(9), and is based on levels estimated to maintain plasma Gpx3 activity at two-thirds of maximal. The issue of optimum Se intake is now further complicated because a number of recent original studies and meta-analyses indicate that Se supplementation may have adverse effects on human health(Reference Duffield-Lillico, Slate and Reid10–Reference Stranges, Marshall, Natarajan, Donahue, Trevisan, Combs, Cappuccio, Ceriello and Reid14). Thus as summarized by Thompson(Reference Thomson15) in a review of assessment tools for Se status and requirements, there is an urgent need to provide further data on which to base reliable estimates of adequate human Se intake as well as on biomarkers for Se status.
Glutathione peroxidase-1 (Gpx1) activity in blood and other tissues has long been known to fall dramatically in Se deficiency in rodents(Reference Hafeman, Sunde and Hoekstra16), in livestock, and other experimental animals. With increasing Se supplementation, this biochemical marker reaches a well-established plateau and has been used to set dietary requirements in experimental animals(17). In addition Gpx1 mRNA levels is also highly regulated by Se in rodents, reach defined plateaus with increasing Se supplementation, and also have been used to evaluate Se requirements(Reference Lei, Evenson, Thompson and Sunde18–Reference Weiss, Evenson, Thompson and Sunde20). In contrast, mRNA level for several of the other well-characterized selenoproteins, including selenoprotein P (Sepp1), glutathione peroxidase-4 (Gpx4), and thyroxine 5′-deiodinase-1 are not well-regulated by Se status in rodents(Reference Lei, Evenson, Thompson and Sunde18–Reference Sachdev and Sunde22). Study of Gpx1 mRNA level has also helped unravel changes in Gpx1 activity during pregnancy and lactation in rats(Reference Sunde, Evenson, Thompson and Sachdev21), further illustrating the value of molecular biology markers for Se status.
The complete set of selenoproteins encoded in the genomes of humans and a number of other species has now been identified(Reference Kryukov, Castellano, Novoselov, Lobanov, Zehtab, Guigo and Gladyshev23), thus potentially providing additional biochemical markers as well as molecular biology biomarkers for assessment of Se status and Se requirements. To evaluate the potential of these markers, we recently determined the impact of overt Se deficiency on the 24 selenoprotein genes in mice using microarrays, and then characterized the Se regulation of mRNA levels in our well-characterized rat model using quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)(Reference Sunde, Barnes, Raines and Evenson24). This identified 2 additional selenoprotein mRNAs besides Gpx1 mRNA that fall dramatically in Se deficiency and that reach plateau levels with increasing Se supplementation(Reference Sunde, Barnes, Raines and Evenson24). Lastly we determined rat selenoprotein mRNA levels in total RNA isolated from whole blood(Reference Evenson, Wheeler, Blake and Sunde25), and found that several of the selenoprotein mRNAs are expressed in rat blood at levels comparable to liver and other tissues. Importantly, blood Gpx1 mRNA levels in Se-deficient rats decreases to 14 % of levels found in Se-adequate samples(Reference Evenson, Wheeler, Blake and Sunde25). Thus whole blood mRNA may be an additional, less-invasive tool for evaluating Se status and perhaps Se requirements in humans.
The use of molecular biology markers to assess human nutrient status is in its infancy. The advent of rapid molecular assays such as microarrays or qRT-PCR provide tools that are already being used to evaluate health and disease in humans and will soon be used for evaluating nutrition as well(Reference Arab26–Reference Janssens, Gwinn, Bradley, Oostra, van Duijn and Khoury28). In addition, it is possible these parameters would provide a more comprehensive evaluation of Se status by better characterizing a hierarchy of Se status over a range of dietary Se intakes, thus potentially providing a more complete set of tools from which to choose for setting appropriate requirements.
Thus we initiated this current collaboration between the University of Wisconsin USA and the University of Reading UK as an opportunity to measure markers of Se status in a British population that was likely to be at suboptimal Se status, based on recent concerns(Reference Rayman3, Reference Elsom, Sanderson, Hesketh, Jackson, Fairweather-Tait, Akesson, Handy and Arthur4). Our initial hypothesis was that in this UK population we would identify a subset of subjects that were marginal in Se status. We found, however, that initial indicators of Se status including plasma Gpx3 activity and selenoprotein mRNA levels were little different from US. volunteers. Thus we expanded the collaboration into a longitudinal study to evaluate Se status in healthy adult Reading subjects using chemical, biochemical and molecular biomarkers.
Methods
Subjects
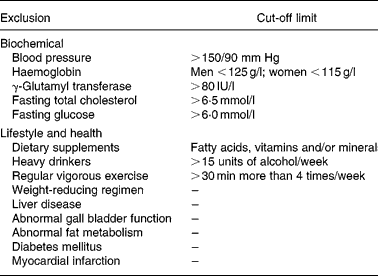
The study was approved by the University of Reading Ethics and Research Committee and the University of Wisconsin Health Sciences Institutional Review Board (IRB protocol: H-2004-0264). This Se study was part of a larger randomized, controlled, crossover dietary study investigating the effect of fruit and vegetable rich concentrated juices(Reference George, Paterson, Waroonphan, Gordon and Lovegrove29). Males and females aged between 30 and 70 years old were recruited from the university and general public in the Reading area. For screening, anthropometric measurements were taken from all subjects and fasting blood samples were sent to the local hospital (Royal Berkshire Hospital, Reading, Berkshire, UK) for analysis of liver function status, lipid concentrations, and fasting glucose. Subjects who did not have any of the exclusion criteria (Table 1) were then randomized with adjustment for age, gender and BMI, to consumption of either the test or control drink intervention.
Table 1 Exclusion criteria and limits
Study design
The main fruit and vegetable study was comprised of thirty-nine volunteers with low fruit and vegetable intake who were randomly assigned to one of 2 groups(Reference George, Paterson, Waroonphan, Gordon and Lovegrove29). After the week 1 run-in period during which subjects consumed 1 portion of cordial (30 ml) diluted with water per day, subjects consumed either fruit and vegetable-rich concentrated juices (100 ml), or 2 portions of a low fruit and vegetable cordial (30 ml) diluted with water per day, for a 6-week period. Test juice selections were carrot and banana, purple carrot and strawberry, or purple carrot and cherry; control intervention choices were 2 portions of Robinson's orange or lemon barley water per day, purchased from local supermarkets. Eight weeks later (week 16) the subjects repeated the 1-week run-in period and then in week 17, subjects repeated the 6-week trial consuming the other intervention drink with the final samples taken in week 23. Mean Se content for test juice consumption was 0·7 μg/d and >0·1 μg/d for the control intervention. For the Se study, fasting plasma was collected at the beginning (weeks 2 or 17) and end (weeks 8 or 23) of the intervention periods. In this longitudinal study, five-day diet records were collected at the beginning and the end of each intervention period.
In addition to the Reading subjects, blood from two US volunteers was obtained and used for method development and standardization, as approved by the University of Wisconsin IRB (H-2004-0264).
Blood analyses
Blood samples were collected from the antecubital vein from all subjects into K2-EDTA vaccutainer tubes (Fisher Scientific, UK), wrapped in foil, and kept on ice for transport to the laboratory. At the laboratory, blood samples were centrifuged immediately after arrival at +4°C at 3000 rpm, and aliquots of plasma were put into separate cryogenic vials for storage for Se and glutathione peroxidase assay. Whole blood samples for ribonuclease protection analysis (RPA) and qRT-PCR were collected in citrated vaccutainer blood tubes (Fisher Scientific, UK), and transported on ice to the laboratory after collection. Under a fume hood, 3 ml fresh whole blood was pipetted into 11·25 ml Tri-Reagent BD (Tri-Reagent BD - RNA, DNA, Protein Isolation Reagent, Molecular Research Center Inc., Cincinnati, OH, USA) in 15 ml polypropylene screw cap conical bottom tissue culture grade centrifuge tubes (Corning Inc., Lowell, MA, USA), and immediately mixed. After 15 min, the tubes were frozen at − 80°C for later shipment to the US.
At the completion of the study, samples for Se, Gpx3 enzyme activity and mRNA determination were shipped on dry ice to University of Wisconsin-Madison Nutritional Sciences Department. On arrival samples, were held at − 80°C prior to analysis. Plasma samples from weeks 2, 8, 17 and 23 of the study from all subjects (n = 39) were analyzed for Gpx3 enzyme activity. Samples were ranked in quartiles based on week 2 Gpx3 activity, and 5 samples from each quartile (20 total) were randomly selected for mRNA determination from weeks 2, 8, 17, and 23 of the study. These sets of 20 plasmas for weeks 2 and 23 were also analyzed for Se.
Plasma Se and glutathione peroxidase (Gpx3)
Plasma Se was analyzed at the University of Missouri (Columbia, MO, USA) using neutron activation as described by McKown & Morris(Reference McKown and Morris30). Plasma Gpx3 activity was analyzed using the method described by Avissar et al. (Reference Avissar, Ornt, Yagil, Horowitz, Watkins, Kerl, Takahashi, Palmer and Cohen31) using 2·0 mmol/l GSH and 0·078 mmol/l tertiary butyl hydroperoxide. For Gpx3 activity, 1 enzyme unit (EU) is the amount of enzyme that will oxidize 1 μmol of GSH per min under these conditions. Protein concentration was determined by the method of Lowry et al. (Reference Lowry, Rosebrough, Farr and Randall32)
RNA isolation and ribonuclease protection analysis (RPA)
Total RNA was isolated from the whole blood-Tri-Reagent BD following the manufacturer's protocol. Yield was ~5–7 μg RNA per ml of whole blood, quantitated using a ND-1000 UV-Vis Spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE, USA).
RPA probe constructs were generated by RT-PCR for Gpx1 from total RNA from male human blood, by PCR-amplification of Gpx4 cDNA from a human heart cDNA library, and using a commercial Gapdh expression vector (pTRI-GAPDH, Ambion Inc., Austin, TX) following standard molecular biology techniques(Reference Ausubel, Brent, Kingston, Moore, Seidman, Smith and Struhl33). Primers were designed to produce the desired size of protected fragment (Gpx1, 538 bp (nt 119-656 of NM_000581); Gpx4, 448 bp (nt 420-867 of NM_001039848); Gapdh, 155 bp (nt 568-722 of NM002046)). RPA probes for Sepp1 and Txnrd1 were also prepared and used, but are not further presented. PCR or RT-PCR fragments were subsequently subcloned into a T-overhang vector (pGEM-T, Promega Corp., Madison, WI, USA), taking advantage of the nonspecific adenosine residues added to the ends of all PCR products. Convenient restriction sites were used in 5′ extensions of each primer to allow ease of subsequent subcloning. Synthesis of 32P-labeled, antisense RNA was then driven from the appropriate promoter.
In vitro transcription of singled-stranded antisense RNA probes was performed according to the manufacturer's protocol (Promega, Madison, WI, USA) as described previously(Reference Sachdev and Sunde22). The specific activity of each probe was modified to balance the intensity of the signals for the different mRNA species within the same gel lane. For example, Gpx1 probe synthesis used 65 μCi of Reference Lowry, Rosebrough, Farr and Randall[32P]UTP (3000 Ci/mmol, NEN, Boston, MA, USA) per reaction, and 2·5 μl of 100 μM cold UTP. Resulting products were gel-purified and ethanol precipitated. For RPA, total RNA samples (10 μg) were heated to 90°C for 15 min and hybridized overnight at 45°C with the single-stranded antisense RNA probes described above, then treated with RNase (40 mg/ml RNase A, 2 mg/ml RNase T1) for 45 min at 30°C. Following RNase inactivation, the protected probe fragments were ethanol precipitated and analyzed on 6 % nondenaturing polyacrylamide gels. Commercial human liver RNA samples (Stratagene, La Jolla, CA, USA) analyzed with each probe individually showed only one major nuclease-protected hybrid fragment, corresponding to the predicted size. Control hybridizations containing all of the probes plus 20 μg of yeast tRNA did not protect any detectable probe fragments from the RNase digestion. Within each sample, the Gpx1 and Gpx4 mRNA signals were normalized to the control mRNA signal (Gapdh).
qRT-PCR analysis
Relative mRNA abundance was determined by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR). RNA (1 μg) was reverse transcribed to cDNA using the RETROscript kit (#AM1710, Ambion), following the manufacturer's protocol. Gene specific primers for the selenoproteins were designed to span a splice-junction and amplify ~150 base segments. Primers used were (gene symbol: 5′ primer no./sequence, 3′ primer no./sequence): hGpx1: 377 gagaatgtggcgtccctct/378 ctcttcgttcttggcgttct; hGpx4: 222 agaccgaagtaaactacactcagc/241 cggcgaactctttgatctct; hSepp1: 246 tcatcaaggaatctcttctcg/227 caagacggccacatctatca; hTxnrd1: 244 tggaactagatggggtctcg/225 cttaactgtctcctcgactttccat; hSelW: 373 cagctcaagaagaagttagaagatga/374 tgaatcaacttcccggctac; hGpx3: 365 gggaggagtacatccccttc/366 ccagaatgaccagaccgaat; hSelH: 381 cttcgaggtgacgctgct/370 cttgaggctcagggaatttg; hGapdh: 249 ggcctccaaggagtaagacc/250 aggggtctacatggcaactg. The final 25 μl qRT-PCR reactions contained 10 ng reverse transcribed RNA, 0·2 mmol/l gene specific forward and reverse primers, and 1X Sybr Green PCR Master Mix (#4309155, Applied Biosystems, Foster City, CA). Reactions were run in the ABI Prism 7000 (Applied Biosystems) with initial stages of 50°C for 2 min and 95°C for 10 min, followed by 50 cycles of 95°C for 15 sec and 60°C for 2 min. A dissociation curve was run for every plate to confirm the production of a single product. The amplification efficiency for each gene was determined from the threshold cycle (C t) values of the samples using the DART program(Reference Peirson, Butler and Foster34). The mRNA relative abundance was calculated according to Pfaffl(Reference Pfaffl35), which accounts for gene-specific efficiencies, and normalizes to the median glyceraldehyde-3-phosphate dehydrogenase (Gapdh) expression set at 100 %.
Dietary analysis
The five-day diet records were analyzed using nutrition analysis software (Diet Cruncher v 1.6, Way Down South Software, Dunedin, NZ) in combination with the electronic version of McCance and Widdowson 6th Edition food composition database (Food Standards Agency (2002) McCance and Widdowson's The Composition of Foods, Sixth summary ed., Cambridge: Royal Society of Chemistry). Where food composition information was not available for a food, data for a generically similar food was substituted. The MAFF Photographic Atlas of Food Portion Sizes (PB 3006, MAFF Publications, London SE99 7TP) was used during subject interviews to improve the quality of the diet records. Because a significant gender effect was observed for Se intake, energy-corrected Se intake (ng Se/kJ/d) was calculated to adjust for the effect of energy consumption on daily Se intake. Effects of gender on markers of Se status were not assessed given the relatively low number of subjects in this study.
Statistical analyses
Statistical analyses were conducted using SPSS v15 (SPSS Inc., Chicago, IL, USA). Data that were not normally distributed following initial descriptive analysis were log transformed and reassessed. Independent t-tests were used to assess plasma Se status between weeks of the study, and 1-way ANOVA was used to assess for significant relationships within the longitudinal part of the study, and within quartiles for each week of the study. Independent t-tests and ANOVA regression modeling was used to assess for relationships between measures. In the figures, the linear regression of the Reading data only is plotted, along with the resulting correlation coefficient (r) and P-value, and in some figures a simple hyperbolic fit (y = ax/(b+x)) of the Reading data is plotted. Significant findings were defined by P < 0·05 at the 95 % confidence interval.
Results
The subjects recruited from the Reading area had a mean age of 45 years and a BMI of 24·5 (Table 2). Analysis of four separate 5-day dietary records over the course of the study found that this population had an average daily Se intake of 48 μg Se/d (range 27–83 μg/d). Males were significantly taller and heavier than females (P < 0·001), and had slight but significantly elevated γ-glutamyl transferase activity (33·5 v. 21·0 IU/l, P = 0·02). Men also consumed significantly more energy than women (P = 0·003) and thus had an average daily Se intake of 54 v. 43 μg Se/d (P = 0·012), respectively. When corrected for differences in energy consumption, however, the daily Se intake of these men and women were 5·9 and 5·8 ng Se/kJ/d, respectively, and no longer significantly different. Biochemical measures other than haemoglobin concentration were not significantly different between men and women, and indicated that this population was in good health.
Table 2 Baseline characteristics and selenium status of subjects

NS = not significant.
* Significance level P < 0·05 by independent t-test.
† Values are mean (sd).
To investigate the potential changes in Se status biomarkers in this longitudinal study, subjects were divided into quartiles based on week 2 values (Tables 3(a) and (b)). There was no significant effect of time on dietary Se intake over the 21 weeks of this study when analyzed for all subjects or by quartile, indicating that there was little change over time in dietary Se intake of these subjects. When dietary Se intake was adjusted for energy intake and expressed as ng Se/kJ/d, there again was no effect of time. Similarly when analyzed by quartile, there was no significant effect of time on plasma Se, plasma Gpx3 activity, or any of the selenoprotein mRNAs, except for the lowest quartile of plasma Gpx3 activity (P = 0·014) and for the highest quartile of Gpx1 mRNA (P = 0·024). Importantly, however, there was no significant effect of time on the averages for plasma Se, plasma Gpx3 activity or any of the selenoprotein mRNAs when analyzed for all subjects. This analysis clearly shows that subjects with low initial levels maintained low levels for each of these biomarkers, those with moderate levels maintained moderate levels, and those with high levels maintained high levels. Thus for each subject the mean of the week 2, 8, 17 and 23 values was used for the remaining analyses.
Table 3a Selenium dietary, chemical and biochemical biomarkers: longitudinal analysis

NS = Not significant.
* Quartiles sorted by week 2 values.
† Significance level P < 0·05 by 1-way ANOVA.
‡ Values are mean (sd).
Table 3b Selenium molecular-biology biomarkers: longitudinal analysis

NS = Not significant.
* Quartiles sorted by week 2 values.
† Significance level P < 0·05 by 1-way ANOVA.
‡ Values are mean (sd).
As the main study was a cross-over design study, the data was analyzed for effect of treatment v. control drink. There was no significant effect of drink-type on any parameter (data not shown).
In these Reading subjects, plasma Gpx3 activity was significantly correlated (r = 0·42, P = 0·007) with dietary Se intake (Fig. 1). When dietary Se intake was adjusted for energy intake, however, plasma Gpx3 activity no longer was significantly correlated (P = 0·17). In addition, the plasma Gpx3 activities found in these Reading subjects were little different from Gpx3 activity levels in plasma from two US volunteers whose blood was used for method development and who had dietary Se intakes of 2·5–2·8 times the mean Se intake of the Reading subjects (Fig. 1). A simple hyperbolic fit for the Reading data only is also plotted in these figures to better illustrate that the plasma Gpx3 values appear to reside on the plateau region of the Se response curves (see discussion).

Fig. 1 Correlations between plasma Gpx3 enzyme activity and dietary Se intake in Reading subjects (●) expressed as μg Se/d (A) or as dietary Se intake adjusted for energy consumption ng Se/kJ/d (B). Values are the means for each subject (n = 39) of values determined at weeks 2, 8, 17 and 23. Shown in each panel is the linear regression line (–), the correlation coefficient and P-value for significance. Also plotted for reference are values from two US volunteers (male: ▾, female: ■) whose blood was assayed at the same time and used for standardization. In addition, a simple hyperbolic fit for the Reading data (…) is also plotted as described in the text.
Plasma Se in these Reading subjects was also significantly correlated with dietary Se intake (r = 0·66, P < 0·002) and this correlation remained significant when adjusted for energy intake (r = 0·52, P = 0·02) (Fig. 2). Again, however, plasma Se concentrations in these Reading subjects were little different from Se concentrations in plasma from the two US volunteers with much higher Se intakes. Lastly, there was no correlation (P = 0·87) of plasma Gpx3 activities with plasma Se concentrations in these Reading subjects (Fig. 3). This suggests that these Reading subjects are on the plateau region of the plasma Gpx3 activity response curve, and this is further emphasized by the data for the two US volunteers which reside on the apparent Se response curve plateaus.

Fig. 2 Correlations between plasma Se concentration and dietary Se intake (●) expressed as μg Se/d (A) or as dietary Se intake adjusted for energy consumption as ng Se/kJ/d (B). Plasma Se values are the means for each subject (n = 20) of values determined at weeks 2 and 23; dietary Se values are means for each subject of values determined at weeks 2, 8, 17 and 23. Shown in each panel is the linear regression line (–), the correlation coefficient and P-value for significance. Also plotted for reference are values from two US volunteers (male: ▾, female: ■), and a simple hyperbolic fit for the Reading data (…).

Fig. 3 Correlation between plasma Gpx3 enzyme activity and plasma Se concentration (●). Plasma Gpx3 values are means for each subject for values determined at weeks 2, 8, 17 and 23; plasma Se values are the means for each subject (n = 20) of values determined at weeks 2 and 23. Shown is the linear regression line (–), the correlation coefficient and P-value for significance. Also plotted for reference are values from two US volunteers (male: ▾, female: ■), and a simple hyperbolic fit for the Reading data (…).
RPA analysis of total RNA from blood from the Reading subjects readily detected mRNA from Gpx1, Gpx4, and Gapdh (Fig. 4), just as we observe in rodents(Reference Sunde, Barnes, Raines and Evenson24, Reference Evenson, Wheeler, Blake and Sunde25). Txnrd1 and Sepp1 mRNA was also readily detected (data not shown). Gpx1 mRNA level normalized to Gapdh levels, however, were not significantly correlated with plasma Se concentrations (P = 0·78), and were little different from Gpx1 mRNA levels found in the US volunteers. Because total RNA yield from human blood sample averages only ~5–7 μg RNA/ml, and because each lane requires 10 μg RNA for the RPA assay, we only conducted limited RPA analysis of the Reading samples, and switched to qRT-PCR analysis for selenoprotein mRNA level.

Fig. 4 RPA analysis for Gpx1 mRNA, Gpx4 mRNA and Gapdh mRNA (A) and correlation between blood Gpx1 mRNA level and plasma Se concentration (B). A: Total RNA (10 μg) isolated from blood from Reading subjects was subjected to RPA analysis. Shown is a representative autoradiogram for RNA isolated from a human male liver (Liver), from blood from 19 Reading subjects, and from blood from male and female US volunteers. Locations of the protected Gpx1, Gpx4 and Gapdh fragments are shown. tRNA lane shows control hybridization reaction with 20 μg of yeast tRNA with all of the probes. B: Resulting Gpx1 mRNA values normalized to Gapdh mRNA levels and set to 100 %, and plotted against plasma Se values (●). Shown is the linear regression line (–), the correlation coefficient and P-value for significance. Also plotted for reference are values from two US volunteers (male: ▾, female: ■) and a simple hyperbolic fit for the Reading data (…).
qRT-PCR for selenoprotein mRNA level in total RNA isolated from human blood revealed that Gpx1 mRNA was 57-fold higher than Gapdh mRNA levels based on Ct values. Relative to Gapdh mRNA, SelH was 3·4-fold, SelW was 0·7-fold, thioredoxin reductase (Txnrd1) was 0·5-fold, Gpx3 was 0·1-fold, Gpx4 was 0·03-fold, and Sepp1 was 0·02-fold of Gapdh mRNA levels. Figure 5 shows the correlation of blood mRNA levels with plasma Se for six selenoproteins, with the mRNA levels normalized to Gapdh mRNA levels in each total mRNA sample. There was no significant correlation between selenoprotein mRNA level relative to plasma Se for Gpx1 (P = 0·36), SelH (P = 0·38), SelW (P = 0·25), Gpx4 (P = 0·06), Gpx3 (P = 0·48), Sepp1 (P = 0·66), or Txnrd1 (P = 0·58, data not shown) mRNA. Dietary Se intake also was not correlated with mRNA level for any selenoprotein mRNA. This lack of correlation reflects the relatively even distribution of lower values across the range of plasma Se values, just as with the selenoprotein enzyme activities, indicating that none of these measures specifically identified individuals with low Se status.

Fig. 5 Correlations between selenoprotein mRNA levels as determined by qRT-PCR and plasma Se concentration (●). Selenoprotein mRNA values are means for each subject for values determined at weeks 2, 8, 17 and 23, normalized to Gapdh mRNA levels and set to 100 %; plasma Se values are the means for each subject (n = 20) for values determined at weeks 2 and 23. Shown is the linear regression line, the correlation coefficient and P-value for significance. Also plotted for reference are values from two US volunteers (male: ▾, female: ■).
Discussion
Gpx1 enzyme activity has been recognised since the early 1970s(Reference Rotruck, Pope, Ganther, Swanson, Hafeman and Hoekstra36) to be highly regulated by Se status; this biomarker was used to establish rodent requirements(17), and the relationship between dietary Se and Gpx1 activity was very helpful in setting initial human dietary Se recommendations(37). Liver Gpx1 activity falls to near zero in Se-deficient rat liver, and rises sigmoidally to a plateau at 0·1 μg Se/g diet, making this a potent biomarker. The discovery that levels of Gpx1 mRNA are also highly regulated by Se status(Reference Saedi, Smith, Frampton, Chambers, Harrison and Sunde38) led to a series of studies in rats showing that Gpx1 mRNA could be used as a molecular biology marker to evaluate Se requirements. Notably, rat liver Gpx1 mRNA levels fall to a low of 6 % of Se-adequate levels, and also rise sigmoidally to a Se response plateau at about 0·05 μg Se/g diet(Reference Lei, Evenson, Thompson and Sunde18–Reference Weiss, Evenson, Thompson and Sunde20). As new selenoproteins were discovered, these studies were confirmed and expanded, such that a hierarchy of dietary Se requirements are observed in the rat, depending on which biomarker is used(Reference Lei, Evenson, Thompson and Sunde18, Reference Sunde, Evenson, Thompson and Sachdev21, Reference Bermano, Nicol, Dyer, Sunde, Beckett, Arthur and Hesketh39–Reference Yang, Hill and Burk41). Use of these molecular biology biomarkers also were most helpful in understanding the Se requirement of pregnant and lactating rats, which exhibit an apparent drop in Gpx1 activity that does not signal an increased dietary Se requirement but rather arises due to a down regulation of steady-state Gpx1 mRNA expression during pregnancy and lactation independent of Se status(Reference Sunde, Evenson, Thompson and Sachdev21). That study in particular emphasizes the need to understand regulation of a biomarker throughout the life cycle, and it illustrates the potential role of molecular biology biomarkers in setting and understanding dietary nutrient requirements.
New biotechnology tools such as qRT-PCR and microarray analysis are making the measurement of molecular biology markers far easier, but practical application in nutrition will require both less invasive sources of RNA than liver for the assays, and require demonstration that these biomarkers can detect individuals and populations that are specifically marginal as well as deficient in a specified nutrient. To make use of our well-characterized rodent model in this expansion, we investigated various sources of mRNA in the rat and found that total RNA isolated from blood has levels of Gpx1 mRNA comparable to those found in liver and other major organs, and we found that Gpx1 mRNA falls to 14 % of Se-adequate levels in Se-deficient rats(Reference Evenson, Wheeler, Blake and Sunde25). A survey of commercially-available total RNA from different tissues revealed that Gpx1 mRNA was readily detected by RPA, and that Gpx1 mRNA in total RNA from human blood was far more predominant than several other mRNAs including Gpx4, Txnrd1, and Sepp1(Reference Wheeler, Evenson and Sunde42, Reference Evenson and Sunde43). This suggested that Gpx1 mRNA in blood had potential as a molecular biology biomarker. Building upon the identification of the complete human selenoproteome(Reference Kryukov, Castellano, Novoselov, Lobanov, Zehtab, Guigo and Gladyshev23), we used microarray analysis to survey the entire selenoproteome in the mouse, and we identified two additional biomarker candidates, SelH and SelW, that were highly regulated in rodents(Reference Sunde, Barnes, Raines and Evenson24), further encouraging us to begin to investigate the potential of molecular biology biomarkers for Se status in humans.
The reports that UK Se intakes had decreased to as low as 29–39 μg Se/d and other reports on related markers(Reference Rayman3, Reference Elsom, Sanderson, Hesketh, Jackson, Fairweather-Tait, Akesson, Handy and Arthur4) suggested to us that UK populations could provide a subset of individuals that were marginal in Se status and thus not on the plateau of the response curves for several Se related biomarkers. Thus we developed a collaboration to evaluate the subjects in the Five-a-Day study(Reference George, Paterson, Waroonphan, Gordon and Lovegrove29) that were being supplemented with control or treatment drinks, each containing only negligible Se, over a 21-week period. Thus, initially, we conceived of this collaboration as a study to evaluate several traditional as well as molecular biology marker in a marginal Se population. Initial analysis of plasma Gpx3 activity, however, revealed that these subjects were little different from the US volunteers who provided blood for method development and standardization. Thus this study became a longitudinal study.
By analyzing 5-day food diaries collected for each subject in weeks 2, 8, 17 and 23, we found that this group of Reading subjects was consuming an average of 48 μg Se/d, which is half of the dietary Se intake of US women and one-third of the Se intake of US men(7). This value matches well with several other recent European studies(Reference Eves and Gesch44–Reference Murphy and Cashman46) but is higher than the 1997 UK report and other reports(Reference Rayman3, Reference Elsom, Sanderson, Hesketh, Jackson, Fairweather-Tait, Akesson, Handy and Arthur4). Plasma Se averaged 1·13 μmol/l in this study and is similar to a number of recent studies(Reference Bates, Prentice, Birch and Delves47–Reference Pagmantidis, Meplan, van Schothorst, Keijer and Hesketh51) but higher than some earlier reports(7, Reference Shortt, Duthie, Robertson, Morrice, Nicol and Arthur45, Reference Bates, Thane, Prentice and Delves52), and considerably higher than concentrations in subjects in Europe and China ( < 0·5 μmol Se/l) where plasma Gpx3 increases sharply with increasing plasma Se(Reference Rannem, Persson-Moschos, Huang, Staun and Akesson53, Reference Xia, Hill and Burk54). This level also lies well-above the initial plasma Se concentration (0·82 μmol/l) in the New Zealand residents from a low Se area that were subjects in the lower half of a screened population(Reference Duffield, Thomson, Hill and Williams55) that participated in one of the two supplementation studies used for setting the 2000 U.S. DRI(56), but this concentration also lies well below the mean US plasma Se concentrations of 1·54 and 1·60 μmol/l for women and men, respectively, of similar ages(7). It is difficult to compare Gpx enzyme activities directly between studies due to differences in assay conditions, especially because Gpx activity is dependent on assay GSH concentration(Reference Flohé, Loschen, Günzler and Eichole57). Plasma Gpx3 activity levels in these Reading subjects, however, were little different from activities concurrently measured in two US volunteers, and also little different from activities we find in an ongoing study on Madison Wisconsin USA subjects (data not shown). This strongly indicates that these Reading subjects are on the plateau region of the Se response curves for plasma Gpx3 activity, plotted as a function of dietary Se, or energy-adjusted dietary Se or plasma Se. This conclusion for a population consuming an average of 48 μg Se/d, however, is consistent with other analyses that concluded that the plateau in plasma Gpx3 activity is reached at an average of 38 μg Se/d in New Zealand subjects and 41 μg Se/d in Chinese subjects in these Se repletion studies(56). For subjects at steady-state for Se, as in these Reading subjects, the required dietary Se intake for plateau levels of Gpx3 activity is likely to be even lower.
Rodent studies allow monitoring of the various biomarkers across the full range of Se status. The resulting sigmoidal or hyperbolic curves clearly show that a number of biomarkers including tissue Se, plasma Gpx3 activity, and tissue Gpx1 activity and mRNA levels can all decrease to < 10 % in Se-deficient rats, and increase to a defined plateau with increasing dietary intake(Reference Lei, Evenson, Thompson and Sunde18–Reference Sunde, Evenson, Thompson and Sachdev21). Similarly, European(Reference Rannem, Persson-Moschos, Huang, Staun and Akesson53) and Chinese(Reference Xia, Hill and Burk54) subjects with plasma Se concentrations < 0·5 μmol Se/l have near zero activity levels of plasma Gpx3 in the most Se-deficient subjects, and show sharply increasing plasma Gpx3 with increasing plasma Se, just as in animals studies, illustrating that these biomarkers do fall in sufficiently Se-deficient human populations. The data in Figs. 1 and 2 suggest that non-plateau levels of plasma Gpx3 or plasma Se only occur in subjects at steady-state ingesting less than ~30 μg Se/d. Thus for Se-replete subjects on the plateau such as in these Reading subjects, traditional biochemical and chemical biomarkers, as well as the molecular biomarkers studied here, do not identify further differences in Se status.
Longitudinal analysis of the Reading subjects who were consuming self-selected diets, and thus presumably at steady-state Se status, indicates that individual subjects changed very little over the 21 weeks. There was no significant effect of time for any Se biomarker in this study. There are few published longitudinal studies that repeatedly analyzed dietary Se intake, plasma Se, and Gpx3 activity in the same a group of subjects over time without Se supplementation. One such study, however, was conducted in 206 Swedish men and women(Reference Akesson, Huang, Persson-Moschos, Marchaluk, Jacobsson and Lindegarde58). At six times over a 12-mo period, dietary Se intake was assessed by 3-day weighed diet records, and plasma Se and Gpx3 activity were measured. Dietary Se intakes in this population were significantly different for men and women, averaging 33 and 26 μg/d, respectively, but were not different when corrected for energy consumption, just as in the present study. Serum Se concentrations averaged 1·10 and 1·17 μmol/l for men and women, respectively, and thus similar to the Reading concentrations of 1·16 and 1·11 μmol/l, respectively, although men were higher than women in the Reading study. Comparison of these two studies shows that the average additional dietary Se intake of +11 μg Se/d for Reading men and +17 μg Se/d for Reading women had a negligible impact on plasma/serum Se concentrations. Thus it is not surprising that there was no significant correlation between plasma Gpx3 activity and energy-corrected dietary Se (P = 0·17) in the Reading study; similarly in the Swedish study, there was no correlation between plasma Gpx3 activity and dietary Se, energy corrected or not. In the present study, there was also no significant correlation between plasma Gpx3 activity and plasma Se, whereas in the Swedish study, this correlation was significant only in men. Lastly, there were no significant effects of time on dietary Se intake and plasma Se concentration in the Swedish longitudinal study; plasma Gpx3 activity was only marginally (~5 %) lower in the first and last sampling periods relative to the intermediate periods only in men(Reference Akesson, Huang, Persson-Moschos, Marchaluk, Jacobsson and Lindegarde58). Thus, overall, both the present Reading study and the Swedish longitudinal study show that Se biomarkers remain relatively constant in populations at steady-state for Se. Furthermore, traditional biomarkers of Se status, plasma Se or plasma Gpx3 activity, were similar and little affected by dietary Se status in both studies, indicating that both populations were on the plateaus of the Se response curves.
This is among the first studies that we are aware of that determined selenoprotein mRNA levels to assess human Se status. In rodents, Se-deficient Gpx1 mRNA levels fall to as low as 6 % of Se-adequate levels in liver(Reference Lei, Evenson, Thompson and Sunde18), and fall to 14 % of Se-adequate levels in total RNA from rat blood(Reference Evenson, Wheeler, Blake and Sunde25). In addition, we have recently found that SelW and SelH mRNA levels in Se-deficient rat liver also fall to < 30 % of Se-adequate levels(Reference Sunde, Barnes, Raines and Evenson24). In total RNA isolated from the Reading subjects, however, there was no correlation between mRNA level and plasma Se concentration for any of the mRNA species that were studied here, including Gpx1, SelW and SelH. Longitudinal analyses of the data further indicated that subjects initially with low mRNA values remained low, and those with high values remained high, indicating that these mRNA measures have potential to identify Se-deficient subjects when applied to a population that is sufficiently deficient. Several recent European human studies have followed changes in selenoprotein mRNA levels following Se supplementation(Reference Meplan, Crosley and Nicol49–Reference Pagmantidis, Meplan, van Schothorst, Keijer and Hesketh51). These study subjects, however, had initial plasma Se concentrations of 1·15–1·19 μmol/l, and thus not unexpectedly no changes with Se supplementation were found for the selenoprotein mRNAs that fall dramatically in Se-deficient rodents, and only a small+11 % change reported for two selenoprotein mRNAs (SelK and Sep15) with 100 μg Se/d supplementation(Reference Pagmantidis, Meplan, van Schothorst, Keijer and Hesketh51).
In summary, assessment of 5-day dietary records at 4 times over a 21 week period found that this Reading UK population had an average daily Se intake of 48 μg Se/d. Daily Se intakes were higher in men than in women, but when adjusted for differences in energy consumption, the daily Se intake of men and women were not different, averaging 5·8 ng Se/kJ/d. Longitudinal analysis indicated that there was little change over time in dietary Se intake of these subjects. Similarly, there was no significant longitudinal effect on plasma Se, plasma Gpx3 activity or any of the selenoprotein mRNAs when analyzed for all subjects. The mean plasma Se concentration was 1·13 μmol/l, which was not significantly different for men and women and similar to concentrations reported in other European studies. Plasma Gpx3 activities, however, were not significantly correlated with energy-adjusted dietary Se intake, nor with plasma Se concentrations, and were not different from levels in US volunteers, all indicating that these Reading UK subjects were on the plateau of the Se response curves. Lastly, RPA and/or qRT-PCR analysis readily detected selenoprotein mRNA for a number of selenoproteins, but none of these potential molecular biomarkers were correlated with plasma Se concentration or energy-corrected dietary Se intake, just as with the selenoprotein enzyme activities. Thus these selenoprotein molecular biomarkers, as well as traditional biochemical markers, so far are unable to distinguish further differences in Se status in these Se replete subjects. Additional biomarkers are needed to characterize the hierarchy of Se status over the full range of dietary Se intakes, and studies will need to be conducted on truly Se-deficient populations to better characterize the potential of selenoprotein mRNA levels as molecular biomarkers for human Se status.
Acknowledgements
This research was supported by the National Institutes of Health (DK74184) for the investigation into novel molecular biomarkers of selenium status, and by Unilever Bestfoods, Heilbronn, Germany, for the research study investigating fruit and vegetable intake.











