Next-generation sequencing (NGS; also known as deep sequencing or ultra-high throughput sequencing) has probably been the most important tool for genomic research over the past few years. NGS has led to numerous discoveries and scientific breakthroughs in the genetic field. The sequencing technology that has entered the research laboratory in the past decade is now being introduced into the clinical diagnostic laboratory. Consequently, NGS results are becoming available in the medical arena as abundance of clinically relevant variants, conferring predisposition to disease, are being discovered at a growing rate (Stanley, Reference Stanley, Sunyaev, Greenblatt and Oetting2014).
Since its first utilization, NGS has been revolutionizing nearly all medical disciplines. Examples include de novo mutation studies (Soler, Reference Soler, Tran-Viet, Galiacy, Limviphuvadh, Klemm, St Germain, Fournié, Guillaud, Maurer-Stroh, Hawthorne, Suarez, Kantelip, Afshari, Creveaux, Luo, Meng, Calvas, Cassagne, Arné, Rozen, Malecaze and Young2013), cancer genomics discovery (Shern, Reference Shern, Chen, Chmielecki, Wei, Patidar, Rosenberg, Ambrogio, Auclair, Wang, Song, Tolman, Hurd, Liao, Zhang, Bogen, Brohl, Sindiri, Catchpoole, Badgett, Getz, Mora, Anderson, Skapek, Barr, Meyerson, Hawkins and Khan2014), prediction of targeted therapy/actionable mutations (Kohsaka, Reference Kohsaka, Shukla, Ameur, Ito, Ng, Wang, Lim, Marchetti, Viale, Pirun, Socci, Qin, Sciot, Bridge, Singer, Meyers, Wexler, Barr, Dogan, Fletcher, Reis-Filho and Ladanyi2014) and noninvasive prenatal testing (Tsui, Reference Tsui, Jiang, Wong, Leung, Chan, Chiu, Sun and Lo2014). The implementation and integration of NGS in these fields is ascertained by the flood of relevant publications (close to 1,500 NCBI PubMed records, under the term ‘next generation sequencing’ or ‘deep sequencing’ in humans, during 2013). This immense inflation of NGS-derived data, its accessibility, its transfer to the medical field and the overall interest means that there is a need for an overview on its implications on the medical arena and the general public.
NGS allows sequencing of an entire human genome (whole-genome sequencing; WGS) or the entire protein-coding DNA, representing 1–3% of the human genome (whole-exome sequencing; WES) in less than a day. This allows for large-scale, comprehensive studies comprising many sequenced individuals to be performed with unprecedented speed.
Decreased sequencing costs (with one major sequencing provider, Illumina, claiming the arrival of the $1000 genome (Hayden, Reference Hayden2014)) have provided remarkable opportunities for the launch of large collaborative projects focused on WGS. As more people have their genome sequenced, the interpretation and understanding of human genetic variance becomes deeper. This in turn facilitates the differentiation between pathogenic variants and variants of uncertain significance. Moreover, this accumulation of population genetic data potentially increases the number of identified clinically relevant variants that are medically actionable.
Certain points have to be considered while debating if genome sequencing should be available to the general public and be performed regardless of clinical indication, whether it is following a physician's recommendation or through a private setting, possibly marketed as an ‘X-ray for the genome’. When facing the decision whether to sequence your own genome, there are several arguments supporting and opposing this decision.
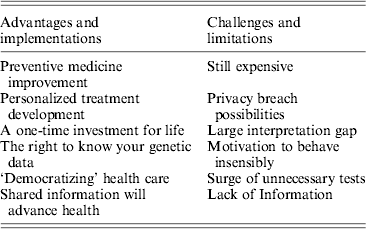
Advantages and implementations:
-
• Development of an effective preventive medicine platform by collecting the entire set of an individual's genetic variations may uncover certain known, disease-associated mutations. Some of these diseases may be alleviated by lifestyle modifications and/or early medical treatment. Early detection of these types of mutations can prevent morbidity and might delay mortality. In a recent report, 42% of people surveyed introduced positive changes into their health behavior after receiving genomics results (Green, Reference Green and Farahany2014). Four pilot research programs were recently launched by the National Institutes of Health (NIH) to explore the potential of newborn genome sequencing in preventing a vast array of disorders and conditions starting at birth (NIH, 2013).
-
• Precision medicine, also known as pharmacogenomics, allows for the elucidation of the effect that specific genetic variants have on the metabolism and action of a certain drug. This field is developing rapidly, with a constant increase in known mutations modulating drug response. The field of personalized medicine was initiated by revealing single nucleotide polymorphisms (SNPs) that account for some of the genetic variability between individuals and made possible the use of genome-wide association studies (GWAS) to examine genetic variation and risk for many common diseases. One example is the clinical impact of the major CYP polymorphisms (CYP2D6, CYP2C19 and CYP2C9) on drug therapy. CYP2D6 is responsible for the oxidative metabolism of about 25% of prescribed drugs (NIH, 2013). The ultra-rapid metabolizer phenotype can cause therapeutic inefficacy of antidepressants, whereas an increased risk of toxicity has been reported in poor metabolizers with several psychotropics (Samer, Reference Samer, Lorenzini, Rollason, Daali and Desmeules2013). Variations in CYP2C19 are associated with decreased clopidogrel metabolic activation, resulting in a decreased antiplatelet effect, and an increased likelihood of a cardiovascular event (Mega, Reference Mega, Close, Wiviott, Shen, Hockett, Brandt, Walker, Antman, Macias, Braunwald and Sabatine2009). Besides SNP detection, NGS enables detection of many other genetic variations – such as small insertions and deletions ranging from 1 to 10,000 bp in length and larger forms of structural variation that may optimize patient drug treatment by tailoring specific pharmacogenomics-based therapy and better monitoring of genomic biomarkers for each individual patient.
-
• While the combined costs of sequencing, data interpretation and storage are still high, they represent a one-time investment for the health provider. Personal sequences remain intact from cradle to grave and a single sequencing effort may aid in countless health events during a person's lifetime.
-
• The public has a fundamental right to know, collect or receive information about their own health, which also includes their genome content, whether it involves positive or negative outcomes. If any government, social or religious agency/group prohibits this privilege, it would require strong legal or moral justification.
-
• ‘Democratizing’ health care, expanding the access given to individuals and enabling the acquirement of as much data as they can in order to make the most informative and founded choices concerning their own health. This follows the historical trend of patient empowerment, a shift from a paternalistic approach to patient care, which tends to ignore personal preferences, towards a patient-centered care in which one's ability to understand and manage his or her own health and disease is improved (No authors listed, 2012). Empowered patients have a better understanding of how to navigate between the many players in the health care system including family, physicians, health insurers and health care regulators. Coping with the complexities of health systems is crucial for achieving better health outcomes. Access to as much information as possible better equips the public to handle this task.
-
• By having an individual's genome sequenced, upon consent, information may be placed in a database, which can be shared by researchers and geneticists around the world. This data can then be used for the development of tools and interpretations based on the genomic information. Unsustainable cost increases threaten the global health care system, and additional progress is stymied more by social than technological factors. Many algorithms have been developed to detect and characterize complex genetic rearrangements, such as insertions, deletions, duplications and inversions, in the human genome. However, these computational tools suffer from relatively high false-positive and false-negative rates compared to SNPs. One suggestion is that larger cohorts are pivotal to improving these success rates. Only by engaging health care consumers (patients) as pioneers who provide both health-related data and insights into pathophysiology can we meet these societal challenges and thus accelerate the pace of biomedical innovation (Hood, Reference Hood and Price2014).
Current challenges and limitations:
-
• Despite the fact that sequencing costs are rapidly declining, the price for computational analysis and data storage remain high, and may pose a financial burden on health care providers and institutions if it becomes a routine procedure.
-
• Although genetic data are considered protected health information under the US Health Insurance Portability and Accountability Act (HIPAA) and similar laws worldwide, many of the protections disappear when the information is ‘de-identified’, meaning that the 18 identifiers specified in the act (including names, addresses, birthdates etc.) are removed (Angrist, Reference Angrist2013). Cross-reference information from public databases to discover the identity of DNA samples donated to research has recently been proven (Gymrek, Reference Gymrek, McGuire, Golan, Halperin and Erlich2013) and might cause any genome-sequenced individual to unintentionally reveal personal information. The risk of re-identification from genomic data sources may affect not only the sequenced individuals, but also their first-degree relatives with whom they share a large portion of their genome sequence. Moreover, we now live in a ‘global village’ – people often move between countries and continents. However there is no global privacy act among the various countries or health providers. While a person might reside in a country with outstanding genetic data protection, they may later move on to countries with no such protections, where leakage of their personal genome data may take place.
-
• A huge gap still exists between the total amount of genomic data resulting from NGS and the interpretable amount according to the current knowledge base. The lack of informed clinicians and geneticists increases this gap, making it challenging for the patient to interpret their own genetic information. The data interpretation gap is demonstrated by the case of 23andMe, a firm offering SNP genotyping services directly to consumers, which was limited by the US Food and Drug Administration (FDA) in December 2013 in order to avoid the pitfalls that can arise from false-positives or false-negatives for certain gene variants. Here we have a ‘catch 22’ situation: this gap cannot be closed unless many thousands of people will contribute their personal genomes along with their health records. The Personal Genome Project (PGP) approach has begun to address this issue by limiting any promises of privacy and confidentiality (Church, Reference Church2005). The project, founded in 2005, is dedicated to creating a shared public platform of public genome, health and trait data. To date, it has more than 2,000 participants, all of whom have agreed to make public, and potentially identifiable, any genomic, medical, environmental and trait data collected about them during the study in order to help science understand the relationship between genotype and phenotype.
-
• Sequencing individuals' DNA and informing them that they have a slightly elevated genetic risk for a certain disease could result in great disservice. This information might motivate them to behave insensibly. This in turn may uncover marketing opportunities for the private sector that will promote all sorts of clinically unnecessary and potentially damaging screening, with further negative and unintended consequences. More than a decade since the Human Genome Project was declared complete, fewer than 60 genetic variants are deemed worthy for use in clinical care, most for severe conditions in very young children (Green, Reference Green, Berg, Grody, Kalia, Korf, Martin, McGuire, Nussbaum, O'Daniel, Ormond, Rehm, Watson, Williams and Biesecker2013). Weeding out irrelevant mutations, focusing on the significant ones and deciding how to act accordingly is a tough mission for health professionals, let alone for the general public.
-
• There would be a surge in demand for all kinds of tests, most of these would be unnecessary and would create a significant burden on the health care system. A recent study claimed that more than a quarter of complex molecular genetic testing orders are unnecessary (Miller, Reference Miller, Krautscheid, Baldwin, Tvrdik, Openshaw, Hart and Lagrave2014) and this number is expected to rise as more patients demand further validations for their genome-sequencing analysis.
-
• Some patients prefer not to know their genetic susceptibilities, especially if there is no cure for these mutated genes. One survey showed that 50–70% of patients would like to be informed about all their results from genomic studies while 16–20% considered genetic information to be riskier than other types of personal medical data (Ruiz-Canela, Reference Ruiz-Canela, Valle-Mansilla and Sulmasy2011). Another study emphasized that the majority of patients desire to have the choice to learn about the probability of conditions that may develop, but given the choice, are not sure how they would act (Townsend, Reference Townsend, Adam, Birch, Lohn, Rousseau and Friedman2012).
-
• The finding that many putatively damaging variants are present as homozygotes in the genomes of healthy individuals raises questions concerning the extent to which these variants contribute to the determination of phenotype. Several mutations reported as ‘disease-causing mutations’ in the Human Gene Mutation Database (HGMD) are likely to be annotation errors (Xue, Reference Xue, Chen, Ayub, Huang, Ball, Mort, Phillips, Shaw, Stenson and Cooper2012). Thus, it has to be carefully considered that NGS projects involving healthy people may result in the incidental discovery of such silent ‘disease-related’ variants (Table 1).
Recently, the American College of Medical Genetics (ACMG) has published a recommendation that laboratories conducting clinical sequencing of patients should report mutations in 56 genes associated with highly penetrant, monogenic disorders regardless of the indication for which the sequencing was ordered, and made the patient's clinician or care team responsible for consulting the patient regarding these results (Green, Reference Green, Berg, Grody, Kalia, Korf, Martin, McGuire, Nussbaum, O'Daniel, Ormond, Rehm, Watson, Williams and Biesecker2013). It is reasonable to speculate that in the foreseeable future, as the cost of such sequencing continues to fall, a majority of health plans will make it easy for their members to have their entire genomes sequenced and linked to their electronic health records for these 56 genes and many others to come. Millions of people will probably be keen to have their genomes sequenced and may even be willing to pay for the sequencing themselves. The challenging part will be, as is today, the clinical interpretation of those genomes (Isakov, Reference Isakov, Perrone and Shomron2013) and the development of useful patient–consumer recommendations. An analogy to radiology, where unexpected anomalies are frequently found, is commonly used to explain this challenging complexity (Solomon, Reference Solomon2014). However, despite impressive ongoing efforts that will continue to yield great progress in the near future, we are not at the point where interpreting a genomic data set is similar to interpreting a radiologic study. It is important not to oversell our abilities to understand and manage the complexity of the human genome in clinical practice.
Though historically human genetic studies focused on the rare and unusual, and was dominated by a handful of experts, the new wave of understanding of the human genome cannot happen without our large-scale participation. Dealing with the inevitable limits of knowledge in a constructive manner is fundamentally different from setting limits to data generation in the presumed ‘best interest’ of the patient or as a method to mask uncertainty and limit the doctor's liability (Lunshof, Reference Lunshof, Church and Prainsack2014; Jarvik, Reference Jarvik, Amendola, Berg, Brothers, Clayton, Chung, Evans, Evans, Fullerton, Gallego, Garrison, Gray, Holm, Kullo, Lehmann, McCarty, Prows, Rehm, Sharp, Salama, Sanderson, Van Driest, Williams, Wolf, Wolf and Burke2014). However, when there is still so much about the function of genes to be uncovered, how useful will free access to DNA sequencing be? And when we reach a point that enough information is gained and interpreted, would you be ready to read your genes in order to know your own medical future?
Table 1. Considerations one should take prior to personal genome sequencing
We thank Dr David Gurwitz, Prof Karen Avraham and Dr Elena Milanesi for commenting on our manuscript.
Funding
The Shomron laboratory is supported by the Israel Cancer Research Fund (ICRF), Research Career Development Award (RCDA); Wolfson Family Charitable Fund; Earlier.org – Friends for an Earlier Breast Cancer Test; Claire and Amedee Maratier Institute for the Study of Blindness and Visual Disorders; I-CORE Program of the Planning and Budgeting Committee, The Israel Science Foundation (grant number 41/11); The Israeli Ministry of Defense, Office of Assistant Minister of Defense for Chemical, Biological, Radiological and Nuclear (CBRN) Defense; Foundation Fighting Blindness; and the Saban Family Foundation – Melanoma Research Alliance.



