Colorectal cancer is the third most common cancer in terms of incidence and mortality in several Western industrialised countries. Thus, every year, nearly one million people worldwide develop colorectal cancer(Reference Jemal, Siegel and Ward1). In the USA, incidence of and mortality from colorectal cancer are estimated at approximately 9 %, accounting for 51 370 estimated deaths and 102 900 new cases per year(Reference Jemal, Siegel and Xu2). In 2006, malignant tumours were the third highest cause of death in Mexico, with colon cancer contributing to 5·0 and 4·6 % for men and women, respectively(3). Proper nutrition is essential for cancer prevention due to the physiological and biological properties of endogenous phytochemical compounds present in the diet. Epidemiological studies have also shown that the importance of diet in the prevention and/or development of cancer was directly associated with the consumption of fruits, vegetables and legumes(Reference Michels4, Reference Millen, Subar and Grawbard5). A high consumption of legumes, such as common beans, has been inversely related to advanced adenoma recurrence in human subjects(Reference Lanza, Hartman and Albert6). Common beans (Phaseolus vulgaris L.) generally contain considerable amounts of non-digestible fraction (NDF) consisting of soluble and insoluble fibres, resistant starch, oligosaccharides, phenolic compounds(Reference Vergara-Castañeda, Guevara-Gonzalez and Ramos-Gómez7) and probably bioactive peptides released in the colon(Reference Torruco-Uco, Chel-Guerrero and Martínez-Ayala8).
The NDF from common bean can be fermented in the large intestine exerting several physiological effects through the production of SCFA, mainly butyrate, propionate, acetate(Reference Campos-Vega, Reynoso-Camacho and Pedraza-Aboytes9, Reference Chen, Ghazawi and Bakkar10), as well as some hydroxyl acids produced from phenolic compounds(Reference Veeriah, Hofmann and Glei11)or by the direct action of compounds that constitute the NDF matrix on several molecular pathways(Reference Torruco-Uco, Chel-Guerrero and Martínez-Ayala8, Reference Kuntz, Kunz and Rudloff12).
Our previous studies(Reference Feregrino-Pérez, Berumen and García-Alcocer13, Reference Campos-Vega, Guevara-Gonzalez and Guevara-Olvera14) demonstrated that the polysaccharide extract or the NDF of common bean modulates gene expression, thereby exerting protection against colon cancer development. A polysaccharide extract of black bean cultivar (cv.) Negro 8025 reduced aberrant crypt foci development in azoxymethane (AOM)-induced rats and regulated the expression of β-catenin, p53, p21, Rb, Bax and caspase-3 (Casp3) genes involved in cell proliferation, cellular arrest and apoptosis(Reference Feregrino-Pérez, Berumen and García-Alcocer13). The NDF from cream bean cv. Bayo Madero, subjected to a simulated monogastric digestive fermentation process, modulated gene expression involved in cell-cycle arrest, induction of apoptosis and proliferation inhibition in an in vitro model of late-stage colon cancer using HT-29 colon adenocarcinoma cells, thus contributing to the chemoprotective effect of common bean against colon cancer development(Reference Campos-Vega, Guevara-Gonzalez and Guevara-Olvera14). The present study investigated the transcriptional effects of the NDF from common bean cv. Bayo Madero on the gene expression profile in the distal colon tissue of Tp53 signal transduction in an in vivo model of early-stage colon cancer, to elucidate the molecular mechanism involved in the chemopreventive action of common bean.
Experimental methods
Materials
Bean cv. Bayo Madero was harvested in 2007 at the Bajio Experimental Station of the National Research Institute for Forestry, Agriculture and Livestock (INIFAP), Celaya, Guanajuato, Mexico. Seeds were cooked using a ‘traditional’ cooking process according to the method of Aparicio-Fernández et al. (Reference Aparicio-Fernández, Manzo-Bonilla and Loarca-Piña15). Male Sprague–Dawley rats were obtained from Harlan, Inc. The care and use of these animals were in compliance with policies and regulations of the Institutional Animal Care and Use Committee of the University of Queretaro, Mexico. AOM was purchased from Sigma Chemical Company.
Non-digestible fraction extraction
The extraction of the NDF was performed following the method of Kurtzman & Halbrook(Reference Kurtzman and Halbrook16). Briefly, water (1·5 litres) was added to 300 g of cooked beans (beans and cooking water) and the mixture was shaken for 1 min and centrifuged (Hermle Z323K; Hermle Labortechnik GmbH) at 9000 g for 10 min. The pellet from the first centrifugation was dissolved in 100 ml of 10 % tannic acid, adjusted to pH 4 and centrifuged again (9000 g for 10 min), and the pellet was washed three times with 100 ml acetone and centrifuged for 10 min after each washing to obtain the NDF. NDF samples were lyophilised and stored in amber flasks at 4°C until further analyses.
Animal and experimental design
Male Sprague–Dawley rats with an initial weight of 69·6 (sd 5) g at 4 weeks of age were used in the present study. The rats were maintained in an air-conditioned animal room at ambient temperature (21 ± 2°C), 55 % humidity and a 12 h light–12 h dark cycle and had free access to a basal diet (2018S Harlan Tekland) and regular tap water. At 1 week after acclimatisation, the rats were randomly placed into two groups (n 12): (1) AOM, basal diet plus subcutaneous injection of AOM (15 mg/kg body weight, dissolved in 1 ml of physiological saline) once per week on weeks 3 and 4; distilled water was also administered intragastrically once per d during the experimental period (9 weeks); (2) NDF from common bean cv. Bayo Madero plus AOM and basal diet (NDF+AOM), basal diet plus AOM (once per week on weeks 3 and 4) and NDF (2·5 g/kg body weight) daily for 9 weeks. The NDF, dissolved in distilled water, was administered intragastrically once per d during the experimental period (9 weeks) and the dose was selected according to the rural per capita intake of beans in the Lagunera Region of Mexico(Reference Del Razo, Garcia-Vargas and Garcia-Salcedo17). The animals were killed 5 weeks after the last injection and the distal colons were removed and stored at − 70°C until analysis. The colon tissues from four rats were randomly chosen for the isolation of RNA. The results on NDF chemoprotection against lesion development of early-stage colon cancer called aberrant crypt foci in Sprague–Dawley rats have been reported by Vergara-Castañeda et al. (Reference Vergara-Castañeda, Guevara-Gonzalez and Ramos-Gómez7).
RNA isolation and complementary DNA synthesis
Total RNA from the distal colon tissue of rats induced with AOM and treated with or without the NDF was isolated using an Rneasy Mini Kit (Qiagen) according to the manufacturer's instructions. All RNA samples were examined for the absence of DNA and RNA degradation by denaturing agarose gel electrophoresis. mRNA (1 μg) was reverse transcribed and amplified with the SMART–PCR complementary DNA (cDNA) synthesis kit and the Advantage cDNA PCR kit (Clontech Laboratories, Inc.). First-strand cDNA synthesis was performed according to the manufacturer's instruction and included 1 μg of total RNA, 7 μl cDNA synthesis (CDS) synthesis primer IIA (12 μm), 7 μl SMART II A oligonucleotide (12 μm) and 200 U Superscript II.
Quantitative RT-PCR (quantitative PCR arrays)
Quantitative determination of Tp53 pathway transcripts was carried out essentially as reported by Campos-Vega et al. (Reference Campos-Vega, Guevara-Gonzalez and Guevara-Olvera14) with slight modifications. Briefly, 46 μl of diluted first-strand cDNA (100 ng/μl) were mixed with the RT2 Real-Time™ SYBR Green/Rox PCR Master Mix (PA-021; SABiosciences). Previously, it was confirmed that 0·5 μl from first-strand cDNA (dilution at 1:1 with free-nuclease water), with the concentration mentioned above, produced the same C t amplification of housekeeping genes included in the PCR array as when using 1 μl. The expression of seventy-seven genes, as a function of the NDF from common bean cv. Bayo Madero treatment, was assessed using the Rat RT2 Profiler real-time PCR array (PARN-027A; SABiosciences), as specified in the manufacturer's user manual. The array included Tp53-related genes involved in apoptosis, cell cycle, cell growth, proliferation and differentiation, and DNA repair plus three housekeeping genes. The quantitative PCR was done using the Strategene Mx 3000P quantitative PCR system (Strategene) with the following protocol: 95°C, 10 min and then forty cycles of 95°C, 15 s/60°C, 1 min. Data were evaluated with MxPro software (Stratagene). The SYBR Green–dsDNA complex signal was normalised to the passive reference dye 6-Carboxyl-X-Rhodamine (ROX), included in the SYBR Green PCR Master Mix to correct for well-to-well fluorescent fluctuations. Relative gene expression levels were calculated by the comparative C t method including normalisation to the constitutively expressed gene and to a control sample. Data were analysed by the PCR array data analysis web portal (http://www.sabioscience.com/pcr/arrayanalysis.php), based on the ΔΔC t method with normalisation of the raw data to either housekeeping genes or an external RNA control. An Excel-based data analysis template was used. We considered sequences as potential target genes if the change between rats treated with NDF+AOM and AOM was greater than 1·1-fold (up- or down-regulated genes; P ≤ 0·05), following the instruction from the data analysis web portal.
Results
Tp53 gene expression pathway analysis showed that seventy-two genes were modulated at least >1·1-fold (induction or inhibition) in the AOM-induced NDF group (NDF+AOM) compared with the AOM group (Tables 1 and 2). These genes belong to different pathways involved in apoptosis, cell cycle, cell proliferation and differentiation, DNA repair and inflammatory response.
Table 1 Up-regulated genes in the colon distal tissue of rats treated with non-digestible fraction (NDF)+azoxymethane (AOM) compared with the AOM group*

* Results were normalised to housekeeping genes, and values represent the degree of changes in mRNA for rats treated with NDF and AOM-induced relative to AOM-induced rats. P ≤ 0·05 compared with the AOM group.
Table 2 Down-regulated genes in the colon distal tissue of rats treated with non-digestible fraction (NDF)+azoxymethane (AOM) compared with the AOM group*

* Results were normalised to housekeeping genes, and values represent the degree of changes in mRNA for rats treated with NDF and AOM-induced relative to AOM-induced rats. P ≤ 0·05 compared with the AOM group.
Tp53, a regulator of different checkpoints during the cell cycle in both G1/S and G2/M phases, was overexpressed (9·3-fold) in the NDF+AOM group compared with the AOM group. In addition, Cdkn1a (p21), participating in the cell-cycle G1/S phase, was also up-regulated (5·5-fold), whereas Ccne2 (Cyclin E) and Cdkn2A were inhibited ( − 2·6- and − 2·4-fold, respectively). Rb1 (retinoblastoma) and E2f1, two important genes involved in this cell-cycle phase, were also suppressed by the NDF treatment ( − 1·5- and − 1·8-fold, respectively). Furthermore, E2f1 and Rb1 can also be suppressed by Dnmt1, overexpressed (9·3-fold) in the NDF+AOM group compared with the AOM group. Myod1 was potently suppressed ( − 11·8-fold).
The NDF+AOM treatment suppressed genes implicated in the G2/M phase of the cell cycle, indicated by a decrease in Ccnb2 and Cdc25c expression ( − 1·4- and − 9·2-fold). Besides, the NDF+AOM group induced Gadd45a expression (18·3-fold) and up-regulated the Sfn gene (6·7-fold), implicated in cell-cycle arrest at the G2/M phase.
The quantitative PCR array also revealed that some genes involved in DNA repair by different mechanisms were regulated in the NDF+AOM group compared with the AOM group. The expression of these genes including Pcna, Msh2 and Xrcc5 increased by 4·6-, 1·8- and 3·2-fold, respectively. Foxo3 can enhance Pcna expression and was up-regulated 2·4-fold by the NDF+AOM treatment.
The genes Bax, Bid and Bnip3 involved in apoptosis were overexpressed in the NDF+AOM group (3·5-, 1·1- and 3·3-fold, respectively), whereas Bcl2, Apaf1, Casp2 and Casp9 were suppressed ( − 3·5-, − 1·2-, − 2·9- and − 1·6-fold, respectively) compared with the AOM group. Moreover, Tp53 induced Stat1 expression (4·2-fold), which enhanced the apoptotic effect against cell damage, and the Ras gene was overexpressed by 5·5-fold in the NDF+AOM group compared with the AOM group. NDF+AOM also regulated the Tp73 l (p63) (up-regulated by 1·3-fold) and Tp73 (down-regulated by − 12-fold) genes.
Nfκb1, Tnf and Traf1 were overexpressed (3·4-, 2·5- and 2·3-fold, respectively) in the NDF+AOM group compared with the AOM group. In the same inflammation process, Il6 and Jun, induced by Nfκb1, were down-regulated by − 1·5- and − 13·7-fold, respectively, in the NDF+AOM group.
We also observed some contrasting and unexpected results, particularly the overexpression of Bag1, Birc5 and Mcl1 (13·7-, 3·7- and 7·3-fold, respectively), and the down-regulation of Rprm ( − 3·3-fold) in the NDF+AOM group compared with the AOM group.
Discussion
The present study shows differential regulation in the expression of several inter-related genes, participating in molecular pathways activated by Tp53 and functioning as a defence stimulus against cell aggression. Their main function is to prevent or delay the development of injuries which could then trigger tumour growth. These results demonstrate the potential of the NDF+AOM treatment in the induced colon tissues to trigger these pathways, and propose the molecular mechanisms preventing the development of colon cancer (Figs. 1–3). AOM was used as a model carcinogenic compound that induced human colon cancer similar to other carcinogens and is an example of non-familial colon cancer in humans. Since the NDF prevented colon cancer induced by AOM in the tested model, the same prevention would be expected from any other carcinogen in humans.
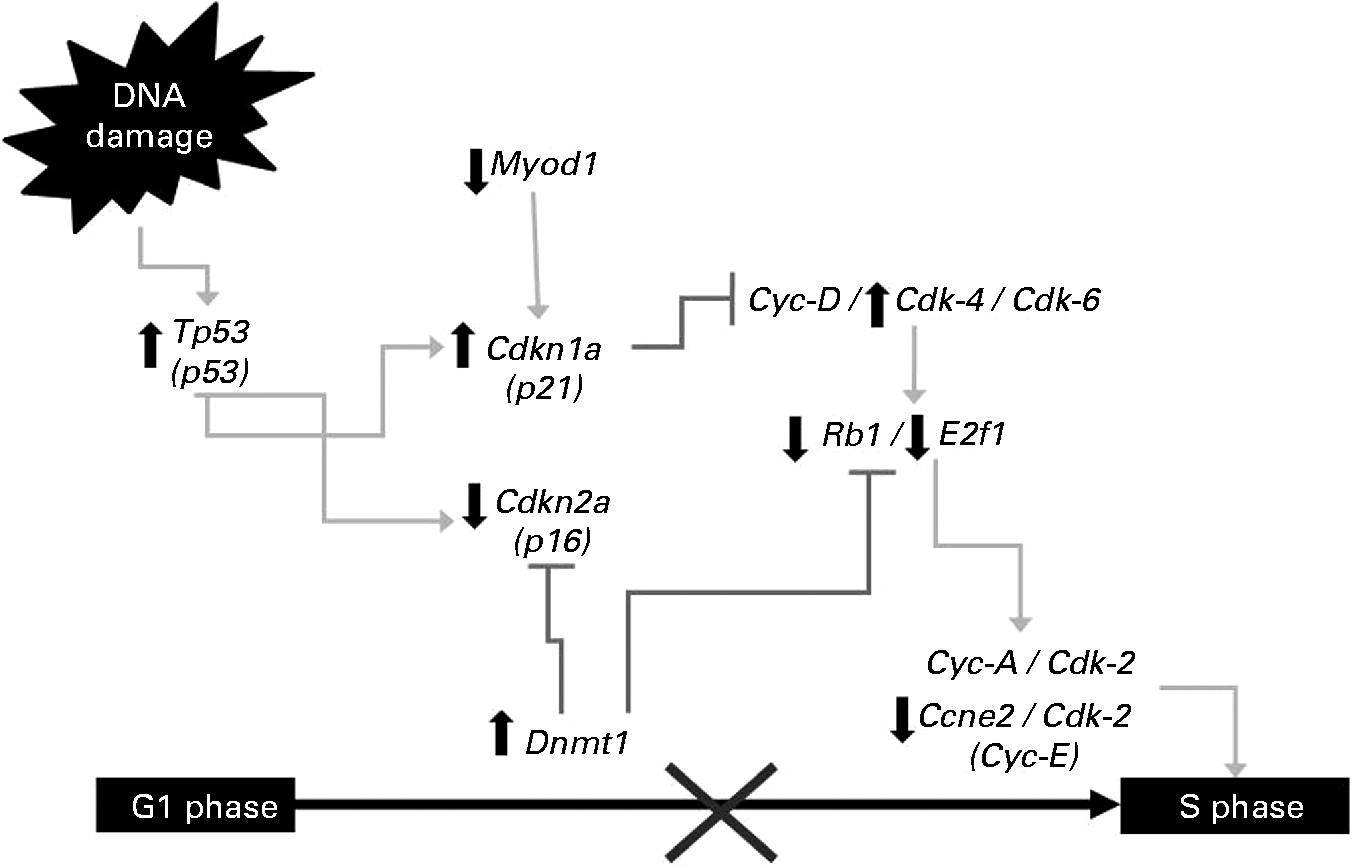
Fig. 1 Changes in gene expression in the G1/S cell-cycle phase. Symbols indicate up-regulation (![]() ) and down-regulation (
) and down-regulation (![]() ) in mRNA expression as derived from array analysis, and signalling pathway interruption (×).
) in mRNA expression as derived from array analysis, and signalling pathway interruption (×).

Fig. 2 Changes in gene expression in the G2/M cell-cycle phase and DNA repair. Symbols indicate up-regulation (![]() ) and down-regulation (
) and down-regulation (![]() ) in mRNA expression as derived from array analysis, and signalling pathway interruption (×).
) in mRNA expression as derived from array analysis, and signalling pathway interruption (×).

Fig. 3 Changes in gene expression in apoptosis and inflammatory pathways. Symbols indicate up-regulation (![]() ) and down-regulation (
) and down-regulation (![]() ) in mRNA expression as derived from array analysis, and signalling pathway interruption (×).
) in mRNA expression as derived from array analysis, and signalling pathway interruption (×).
The tumour-suppressor gene Tp53 activates or suppresses the transcription of target genes involved in the repair of cell injuries in normal conditions or in response to cell stress or genotoxicity(Reference Ho and Benchimol18, Reference Rahman-Roblick, Roblick and Hellman19). The stabilisation and activation of p53 protein is critical in stress response, but since p53 gene expression is rapid and transient, increasing the transcription rate of the gene is crucial. Moreover, induction of p53 mRNA levels increases in parallel with the rate of the newly synthesised p53 protein(Reference Li, Rao and Guo20). In the present study, Tp53 was overexpressed in the NDF+AOM group compared with the AOM treatment. Our previous studies showed no adverse effects due to NDF treatment in rats without AOM(Reference Vergara-Castañeda, Guevara-Gonzalez and Ramos-Gómez7). Moreover, Feregrino-Pérez et al. (Reference Feregrino-Pérez, Berumen and García-Alcocer13) reported that common beans (Phaseolus vulgaris L.) cv. Negro 8025 without AOM did not induce p53 expression, therefore the modulation of p53 expression by the NDF treatment may be considered as part of the response to carcinogen and non-NDF. Several genes, such as Cdkn1a (p21) acting as Tp53 transcriptional targets leading to a first control point in the G1 phase of the cell cycle(Reference Mahyar-Roemer and Roemer21), were up-regulated, whereas Ccne2 (Cyclin E) and Cdkn2a were inhibited by the NDF treatment (Fig. 1). Once Tp53 induces p21 transcription, it inhibits the cyclin–Cdk complex necessary for the G1-to-S-phase(Reference Damia and Broggini22) and G2-to-M-phase transitions in colon cancer cells(Reference Maeda, Chong and Espino23).
The retinoblastoma gene (Rb1) encodes a 105 kDa nuclear phosphoprotein, which in the non-phosphorylated state can bind and suppress the E2f1 transcriptional factor, essential for the G1-to-S-phase transition(Reference Foijer and Te Riele24). Both genes were down-regulated by the NDF treatment compared with the AOM group. The p21, Ccne2, Rb1 and E2f1 regulation suggests a possible cell-cycle arrest in the G1/S phase induced by the NDF treatment (Fig. 1). The change in p21 and Rb1 expression is consistent with cell-cycle arrest at the G1 phase in the distal colon of AOM-induced rats by the treatment of a polysaccharide extract obtained from black bean cv. Negro 8025, reported by Feregrino-Perez et al. (Reference Feregrino-Pérez, Berumen and García-Alcocer13). On the other hand, oligosaccharides have been reported to induce cell-cycle arrest in different cell lines of human colon cancer through the regulation of p21, cyclins and some kinase expression(Reference Kuntz, Kunz and Rudloff12). The NDF treatment contains considerable amounts of oligosaccharides (raffinose, stachyose and verbascose) quantified by HPLC(Reference Vergara-Castañeda, Guevara-Gonzalez and Ramos-Gómez7), and these compounds probably influence cell-cycle arrest by modulating these genes.
Moreover, another methyltransferase gene, Dnmt1, overexpressed in the NDF+AOM group compared with the AOM group (Fig. 1), was able to inhibit gene transcription involved in proliferation and cell-cycle progression(Reference Robertson, Ait-Si-Ali and Yokochi25). Cdkn2a suppressed in this study also suggest Dnmt1 association with gene silencing by DNA methylation for the gene promoter regions of Cdkn2a (Reference Robert, Morin and Beaulieu26). Furthermore, the Myod1 gene participating in apoptosis and cell differentiation(Reference Harford, Shaltouki and Weyman27) was also suppressed, thereby demonstrating that it could be highly methylated and therefore silenced(Reference Kawakami, Ruszkiewicz and Bennett28, Reference Hiranuma, Kawakami and Oyama29).
Tp53 transcriptionally supresses key regulators such as Cdc25c and Ccnb2, cyclin that complexes with Cdc2 to induce mitosis, and this inhibition promotes cell-cycle arrest before the cell enters mitosis(Reference Imbriano, Gurtner and Cocchiarella30). In the present study, Ccnb2 and Cdc25c were suppressed, probably resulting in cell-cycle arrest at the G2/M phase (Fig. 2). Gadd45a gene transcription also activated by Tp53 acts as a control point in the G2/M transition of the cell cycle, contributing to Ccnb2 inhibition(Reference Hildesheim and Fornace31). Gadd45a has also been implicated in excision DNA repair through the interaction with proliferation cell nuclear antigen (Pcna)(Reference Jung, Kim and Mun32). This gene was overexpressed by the NDF treatment, suggesting that both overexpression and interaction confer important protective mechanisms against aberrant crypt foci development involving DNA repair. Pcna interacts with Msh2 at the early stages of the DNA repair process by recognising loss bases(Reference Li, Liu and Wang33), and it was up-regulated by the NDF. In addition, the NDF also induced Xrcc5, a gene involved in repairing the double-strand break DNA molecule by directly binding to DNA and recruiting other repair proteins(Reference Sengupta and Roychoudhury34). These results support the potential induction of DNA repair by the NDF due to synergism among different genes mentioned as an alternative protective mechanism.
The product of the Sfn gene (14-3-3-σ), implicated in cell-cycle arrest between the G2 and M phase(Reference Hermeking, Lengauer and Polyak35), can bind and inhibit several cyclin-dependent kinases (Cdk2, Cdc2 and Cdk4) and Cyclin B1 and 2(Reference Laronga, Yang and Neal36), and inactivate Cdc25c, which is inhibited by the NDF, preventing mitosis initiation. Sfn was induced by the NDF (Fig. 2), suggesting that modulation of this gene could contribute to cell-cycle arrest in the G2/M phase through Ccnb2, Cdc25c and Gadd45a (Reference Luk, Siu and Lai37).
The Foxo3 gene, mediating cell proliferation, survival, differentiation, DNA repair and defence against oxidative stress(Reference Lam, Francis and Petkovic38), was enhanced by the NDF (Fig. 2). Foxo3 induces Gadd45a, Cdkn1a and Bnip3 transcription(Reference Lam, Francis and Petkovic38, Reference Greer and Brunet39), which suggests that overexpression of these genes in the present study may also be induced through the direct action of Foxo3, functioning as a transcription factor or promoting the activity and stability of Tp53, as reported in other studies(Reference You, Yamamoto and Mak40).
Ras can lead the process normally associated with the acquisition of a transformed phenotype or promote growth detention by cell-cycle arrest and cell death by apoptosis. The NDF increased Ras expression compared with the AOM group (Fig. 3), presumably to induce Cdkn1a (p21) expression and decrease Cdc25 mediated by Ras (Reference Frame and Balmain41).
Tp53 is also a key transcription factor inducing apoptosis by modulating several genes involved directly or indirectly in molecular pathways resulting in programmed cell death(Reference O'Brate and Giannakakou42). The NDF induced various genes involved in apoptosis. The apoptosis intrinsic pathway was modulated by the pro-apoptotic genes Bax and Bnip3, which in turn suppress the anti-apoptotic gene Bcl2 (Fig. 3). The Bax:Bcl2 ratio determines the susceptibility of a cell to die by apoptosis by mitochondrial membrane depolarisation(Reference Pawlowski and Kraft43, Reference Bai and Meng44), and Bnip3 directly inhibits Bcl2 after activation by intracellular death signals(Reference Ray, Chen and Velde45).
Once cytochrome c is liberated into the cytosol from the mitochondria, it forms a complex with Apaf1 and Casp9 called apoptosome, which activates Casp3, an enzyme responsible for DNA fragmentation and cell death by apoptosis(Reference Chen, Willis and Wei46). In the present study, Apaf1, Casp2 and Casp9 were down-regulated, suggesting that apoptosis induction can be carried out by cytochrome c but independent of Casp2 and Casp9 activation, as evidenced by Marsden et al. (Reference Marsden, Ekert and Van Delft47). Casp2 and Casp9 are not essential to induce apoptosis in thymocytes 2− / −9− / − cell, since the death process presented the same characteristics of apoptosis and probably death involved the action of other caspases. We suggest that apoptosis is triggered by Casp3 activation directly through cytochrome c or by a previously proposed caspase-independent path(Reference Shrivastava, Tiwari and Sinha48). Shrivastava et al. (Reference Shrivastava, Tiwari and Sinha48) suggested that iodine-induced apoptosis on MCF-7 cells (cells without a functional caspase-3 expression) is independent of caspase activation and involves the loss of membrane polarity, increases Bax expression, decreases Bcl2 expression and releases an apoptosis-inducing factor from the mitochondrial membrane. The release of an apoptosis-inducing factor, Smac/DIABLO, HtrA2/Omi or Endo G, from the mitochondrial membrane induces cell death independently of caspases(Reference Ravagnan, Roumier and Kroemer49).
Bax induction and Bcl2 inhibition by the NDF of common bean cv. Bayo Madero are in agreement with the effect of polysaccharides of bean cv. Negro 8025 in AOM-induced rats observed by Feregrino-Perez et al. (Reference Feregrino-Pérez, Berumen and García-Alcocer13). Moreover, the NDF of Bayo Madero induced Bid expression, suggesting that this gene could activate other caspases (Fig. 3). Modulation of the Bax, Bnip3, Bid and Bcl2 genes suggests a potential effect of the NDF on cell death activation by apoptosis. Yu et al. (Reference Yu, Li and Liu50) showed that Bax mRNA was overexpressed and Bcl2 repressed in HT-29 cells by genistein, an isoflavone, in a dose-dependent manner. The NDF of Bayo Madero contains phenolic compounds and condensed tannins(Reference Vergara-Castañeda, Guevara-Gonzalez and Ramos-Gómez7) that can partly reach the colon intact and exert protective effects on the cell in that organ.
Another pathway to promote apoptosis could be induced by Stat1 that was overexpressed in the NDF+AOM group (Fig. 3). Stat1 is an important gene that optimally triggers apoptosis by multiple stimuli through p21 induction, involving cytochrome c release and Casp3 activation(Reference Agrawal, Agarwal and Chatterjee-Kishore51).
Gene induction of different molecular pathways can also be modulated by two other members, Tp73 l (p63) and Tp73, of Tp53 superfamily transcription factors, whose functions are similar, but not identical, to Tp53 (Reference Lin, Sengupta and Gurdziel52). Tp73 l and Tp73 were up- and down-regulated, respectively (Fig. 3), suggesting that at least p63 (overexpressed) contributed to the induction of DNA repair genes, enhancing the apoptotic effects of Tp53 (Reference Flores, Tsai and Crowley53) resulting in chemoprotection by the NDF from common beans.
Pten is involved in cell adhesion, migration and invasion by inhibiting the adapter protein Shc and the kinase protein of focal adhesion Fak (Reference Haier and Nicolson54). In the present study, the NDF induced Pten expression (Fig. 3), suggesting that its chemopreventive effect could be promoted by avoiding cell signalling, and the damage generated by AOM was not extended to other cells.
Nfκb1 is a key gene in the innate inflammatory response and cell survival. However, this gene has a paradoxical role because it also exerts a pro-apoptotic function, under certain circumstances, through the induction of some genes such as Tnf death receptors(Reference Campbell, Rocha and Perkins55) and Tp53 expression and stabilisation by initiating the apoptosis signalling cascade(Reference Fujioka, Schmidt and Sclabas56) (Fig. 3). The NDF induced Nfκb1 and Tnf expression in AOM-induced rats, indicating the contribution of Nfκb1 to trigger apoptosis through Tnf death receptors indirectly by Tp53 induction. These data suggest that both mechanisms could be activated by Nfκb1 at the early stage of colon cancer and the gene probably has not yet suffered any mutations or aberrations that block its pro-apoptotic activity, becoming a potential anti-apoptotic function, as suggested by Wu & Miyamoto(Reference Wu and Miyamoto57).
Jun antagonises the pro-apoptotic and anti-proliferative activity of Tp53 in the initiation stage of cancer development(Reference Maeda and Karin58). The phosphorylation of Jun by c-Jun N-terminal kinase leads to the activation of the Il6 gene(Reference Cahill and Rogers59), which is normally induced in the inflammatory response(Reference Ahn and Aggarwal60). The NDF suppressed Jun and Il6 in the present study (Fig. 3), indicating that the NDF treatment presumably protects against an inflammatory response by inhibiting the pro-inflammatory pathway mediated by Il6.
Tissue response to aggression caused by AOM and the protection provided by the NDF from common bean cv. Bayo Madero also showed some contradictions. Examples of such events are as follows: Bag1 overexpression, an important gene for tumour growth and progression(Reference Clemo, Collard and Southern61); Birc5 (survivin) induction, an important inhibitor of apoptosis and a proliferation promoter in colorectal cancer(Reference Chen, Liu and Fu62); Rprm inhibition, a gene that induces cell-cycle arrest between the G2 and M phase, regulating Cdc2 and Cyclin B1 activity(Reference Ohki, Nemoto and Murasawa63). The activation of contradictory events of signalling pathways and the dynamic balance between them may be important for cell survival or apoptosis. This issue is a matter of each individual cell, since each cell responds to damage and achieves a physiological state by either apoptosis or survival(Reference Nair, Yuen and Olanow64, Reference Iacomino, Tecce and Grimaldi65). Moreover, the unexpected overexpression of oncogenes and the decreased expression of tumour-suppressor genes may also reflect the analysis of different cell types along the crypts. In a normal colon, morphogenesis genes involved in cell cycle and proliferation are mainly expressed at the crypt base, whereas apoptosis-inducing genes are expressed at the crypt top, and the results obtained from the PCR array represent the sum of gene expression along the crypt(Reference Kosinski, Li and Chan66).
In conclusion, the present study describes changes in gene expression profile in the distal colon tissue of AOM-induced rats in response to treatment with the NDF of common bean cv. Bayo Madero at an early stage of colon cancer. Additionally, the present study proposes the scientific basis by which the NDF has a chemopreventive effect against colon cancer development through modulating different molecular mechanisms such as apoptosis induction, cell-cycle arrest, inhibition of cell proliferation and inflammation and induction of DNA repair (Figs. 1–3).
Acknowledgements
The present study was supported by the Consejo Nacional de Ciencia y Tecnologia (CONACYT), grant no. 57536. We thank Virginia Dickison for technical support with quantitative PCR arrays and the Instituto Nacional de Investigaciones Forestales Agricolas y Pecuarias, the Bajio Station (INIFAP, Celaya). V.-C. H. designed and conducted the research, analysed the data and wrote the paper; G.-G. R. provided technical support and supplied material for RNA extraction, cDNA synthesis and PCR arrays; G.-O. L. provided technical support and supplied material for RNA extraction and cDNA synthesis; O. B. D. supplied material for PCR arrays; R.-C. R. provided technical support; W. P. provided technical support and supplied material for PCR arrays; L.-P. G. designed and supervised the trial as principal investigator, and participated in drafting and revising the manuscript. All authors contributed to the data interpretation and approved the final version of the manuscript. There are no conflicts of interest.







