INTRODUCTION
The burden and severity of typhoid fever infections caused by Salmonella enterica serovar Typhi is a significant worldwide public health concern, particularly in developing countries. Typhoid fever is endemic in Asia (particularly south-central and south-east Asia), Africa, Latin America, the Caribbean, and Oceania (excluding Australia and New Zealand), with an estimated global burden of 21 million illnesses and 216 000 deaths each year [Reference Crump, Luby and Mintz1].
Humans are the only natural reservoir of S. Typhi, with transmission occurring through consumption of faecally contaminated food and water [Reference Basnyat2–Reference Kubota5]. S. Typhi infections may cause bacteraemia, fever, headache, malaise, abdominal distress, diarrhoea, and ‘rose spot’ rash and about 3–5% of those infected progress to a chronic carrier state [Reference Basnyat2, Reference Kubota5, Reference Guzman6]. The majority of cases in endemic areas are in children aged 5–19 years and mortality rates can reach up to ⩾10% [Reference Bhan, Bahl and Bhatnagar3, Reference Brachman7–10]. Incidence of typhoid fever is low in developed countries, although sporadic infections associated through contact with chronic carriers, food handlers or acquired during travel to endemic regions place a burden and significant costs on public health systems [Reference Bhan, Bahl and Bhatnagar3, Reference Kubota5]. In the USA, the exposure to chronic carriers is estimated to cause up to 30% of all infections [Reference Bhan, Bahl and Bhatnagar3], whereas travel-associated infections, primarily to the Indian subcontinent, have risen from 33% in 1972 to 81% in 1996 [Reference Ackers11].
Antimicrobial-resistant (AMR) typhoid strains have emerged, and there is particular concern for those strains resistant to first-line antimicrobials typically used for treating these infections. In endemic areas there is now widespread resistance to multiple first-line antimicrobials such as ampicillin, chloramphenicol, streptomycin, sulfadiazine, tetracycline and trimethoprim [Reference Kubota5, Reference Pang8, Reference Hampton12]. Multidrug-resistant (MDR) S. Typhi strains that are resistant to at least ampicillin, chloramphenicol and trimethoprim–sulfamethoxazole are now the predominant epidemic clone in South Asia and the Indian subcontinent; travel to these regions has been identified as a significant risk factor in acquiring MDR S. Typhi infections [Reference Kubota5]. Fluoroquinolones and some third-generation cephalosporins are currently the antimicrobials of choice for treating infections acquired in these endemic regions [Reference Ackers11, Reference Kumar, Rizvi and Berry13]. High rates of nalidixic acid resistance (NAL-R) have been reported in strains [Reference Pang8, Reference Ackers11], raising concerns about increased risks for the emergence of fluoroquinolone and cephalosporin resistance [10, Reference Ackers11, Reference Kumar, Rizvi and Berry13–Reference Crump15]. Patients infected with NAL-R strains require higher fluoroquinolone dosages and longer, more expensive treatments. They also tend to experience higher stool carriage rates and higher case-fatality rates [Reference Connor and Schwartz4, 10, Reference Ackers11]. An increase in fluoroquinolone resistance in S. Typhi strains will limit the value of the use of this class of antibiotic for empirical treatment in the future.
Pulsed-field gel electrophoresis (PFGE) is an effective tool to subtype S. Typhi for surveillance purposes; to identify and investigate outbreaks, as well as to characterize endemic strains [Reference Ben Saida16–Reference Salve19]. While S. Typhi isolates from the same geographic region may not have similar PFGE patterns [Reference Kubota5], some countries do report single predominant patterns [Reference Salve19]. Current evidence also suggests that MDR strains are genetically very similar and that PFGE may be useful in identifying these strains [Reference Kubota5, Reference Kam17, Reference Thong, Bhutta and Pang20]. In addition, since the 1950s, phage typing has been extensively used worldwide to characterize S. Typhi [Reference Anderson and Williams21–Reference Navarro25].
We characterized S. Typhi strains collected from 2000 to 2006 through national laboratory-based surveillance in Canada by PFGE, phage typing and antimicrobial resistance determination. Establishing infection rates, defining the clinical epidemiology, and describing the diversity of confirmed typhoid strains in Canada can be used to evaluate current treatment practices as well as to identify the emergence of strains with novel characteristics and will contribute to the understanding of the global dissemination of S. Typhi.
MATERIALS AND METHODS
Bacterial isolate identification and characterization
Identification and serotyping
Bacterial isolations and presumptive identifications from clinical specimens were performed in local clinical laboratories and forwarded to the provincial public health and central reference laboratories for identification and serotyping according to the National Enteric Surveillance Program (NESP) of the Public Health Agency of Canada [26]. Strains with incomplete identification were forwarded and confirmed at the National Microbiology Laboratory (NML) using the serotyping methods described by Ewing [Reference Ewing27] and Popoff [Reference Popoff28]. Forwarded isolates were accompanied by a minimal amount of case information (age, gender, collection date, and province).
Phage typing
All isolates were phage typed at the NML as described previously [Reference Anderson and Williams21, Reference Adams29, Reference Farmer, Hickman and Sikes30] using S. Typhi Vi typing bacteriophage prepared and supplied by the Centre for Infections, Laboratory of Enteric Pathogens, UK.
PFGE
A total of 222 isolates were characterized by PFGE (114 isolates from 2004, 101 isolates from 2005 plus seven other isolates from 2003 and 2006) via the PulseNet Canada Network. PFGE using enzyme XbaI was performed by either the provincial laboratory or at the NML following internationally standardized PulseNet methods [Reference Barrett31–33]. During the time-frame of this study, PFGE was not routinely performed on all S. Typhi isolates. To obtain a representative PFGE dataset, a subset of isolates from 2004 and 2005 was selected for further study. All PFGE patterns were compared to the national PulseNet Canada database and assigned pattern names/designations according to standardized PulseNet methods [34] using BioNumerics version 4.5 (Applied Maths, Belgium). Cluster analysis to determine PFGE groupings was performed using the Unweighted Pair Group Method with Arithmetic Mean and the Dice coefficient, 1·5% optimization, 1·5% tolerance.
Antimicrobial susceptibility testing
AMR profiles were determined for all isolates. Isolates collected from 2000 to 2002 were tested using the Kirby–Bauer disc diffusion technique [Reference Bauer35] using BBL Sensi-Disc antimicrobial susceptibility discs (Becton Dickinson, USA) according to Clinical Laboratory Standards Institute (CLSI) guidelines [36, 37]. Antibiotic discs employed were ampicillin (AMP) at 10 μg, chloramphenicol (CHL) at 30 μg, ciprofloxacin (CIP) at 5 μg, streptomycin (STR) at 10 μg, sulfamethoxazole (SMX) at 0·25 μg, tetracycline (TCY) at 30 μg, and trimethoprim–sulfamethoxazole (SXT) at 1·25/23·75 μg. Since 2003, all S. Typhi isolated in Canada by the provincial public health laboratories and reference centres have been forwarded to the NML as part of the Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) program and tested via broth microdilution methods using the Sensititre Automated Microbiology System (Trek Diagnostic Systems, USA). Fifteen antimicrobials are included in the susceptibility panel, including: amikacin (AMK), amoxicillin/clavulanic acid (AMC), AMP, cefoxitin (FOX), ceftriaxone (CRO), ceftiofur (TIO), CHL, CIP, gentamicin (GEN), kanamycin (KAN), nalidixic acid (NAL), STR, sulfisoxazole (SSS), TCY, and SXT. The resistance breakpoints were those derived from CLSI [37, 38] and intermediate resistances were considered susceptible. For analysis, SMX and SSS susceptibilities are combined as sulfonamide (SUL) susceptibility. MDR is defined as resistance to three or more classes of antibiotics based on mechanism of action, that include: aminoglycoside (AMK, GEN, KAN, STR); β-lactam (AMC, AMP, FOX, CRO, TIO); chloramphenicol (CHL); quinolone (NAL, CIP); sulfonamide (SMX, SSS, SUL); tetracycline (TCY); trimethoprim–sulfamethoxazole (SXT).
Epidemiological data collection and statistical analysis
Each isolate of S. Typhi was assumed to represent an individual case of typhoid fever. Ages were grouped into the following categories used under the National Studies for Acute Gastrointestinal Illness (NSAGI): 0–12, 13–17, 18–24, 25–54, 55–64, and ⩾65 years. A request was sent to provinces that reported typhoid cases occurring during 2005 and 2006 to extract further epidemiological data from their existing databases for all cases investigated or reported during this time period. The variables of province, age, gender, and travel history were captured for analysis in this study. Additional epidemiological information was extracted by provincial epidemiologists and reported in Excel (Microsoft, USA). Data were analysed using SAS version 9.2 (SAS Institute Inc., USA). Statistics Canada yearly population projections were used as the denominator in calculating age- and gender-specific isolation rates/100 000 inhabitant-years [39]. Differences in seasonality were investigated using a χ2 test, with seasons defined as follows: December–February (winter), March–May (spring), June–August (summer), and September–November (autumn). Data collected using the International Travel Survey were used to calculate incidence of notified infections by country/100 000 Canadian resident travellers [40]. Multivariate analysis using proc logistic (SAS version 9.2; SAS Institute Inc., USA) was conducted to compare the presence of antimicrobial resistance across years and provinces. Odds ratios were obtained using the logistic procedure.
RESULTS
Descriptive epidemiology of S. Typhi infections
A total of 880 laboratory-confirmed S. Typhi isolations were reported in Canada from 2000 to 2006, with a total number of isolates increasing from 1% (89/6297) to 3% (178/5870) of all Salmonella reported. The isolation rate also increased from 0·29 in 2000 to 0·55 isolations/100 000 inhabitants in 2006 (Fig. 1). The number of S. Typhi cases per region varied from no cases in the northern territories to a total of 501 in the province of Ontario over the 7-year period. British Columbia had the highest isolation rate of S. Typhi, as well as the greatest increase in rates between 2000 (0·37/100 000 inhabitants) and 2006 (1·02/100 000 inhabitants), followed by Ontario, Alberta, and Québec (Fig. 1). The number of isolates identified peaked during the months of March and September (Fig. 2) with the lowest during November. There was a significant seasonal variation in the isolation of S. Typhi (χ2=23·50, d.f.=3, P<0·0001), with 31% of the isolates identified during spring and 26% in summer, followed by 24% in autumn and 19% in the winter.
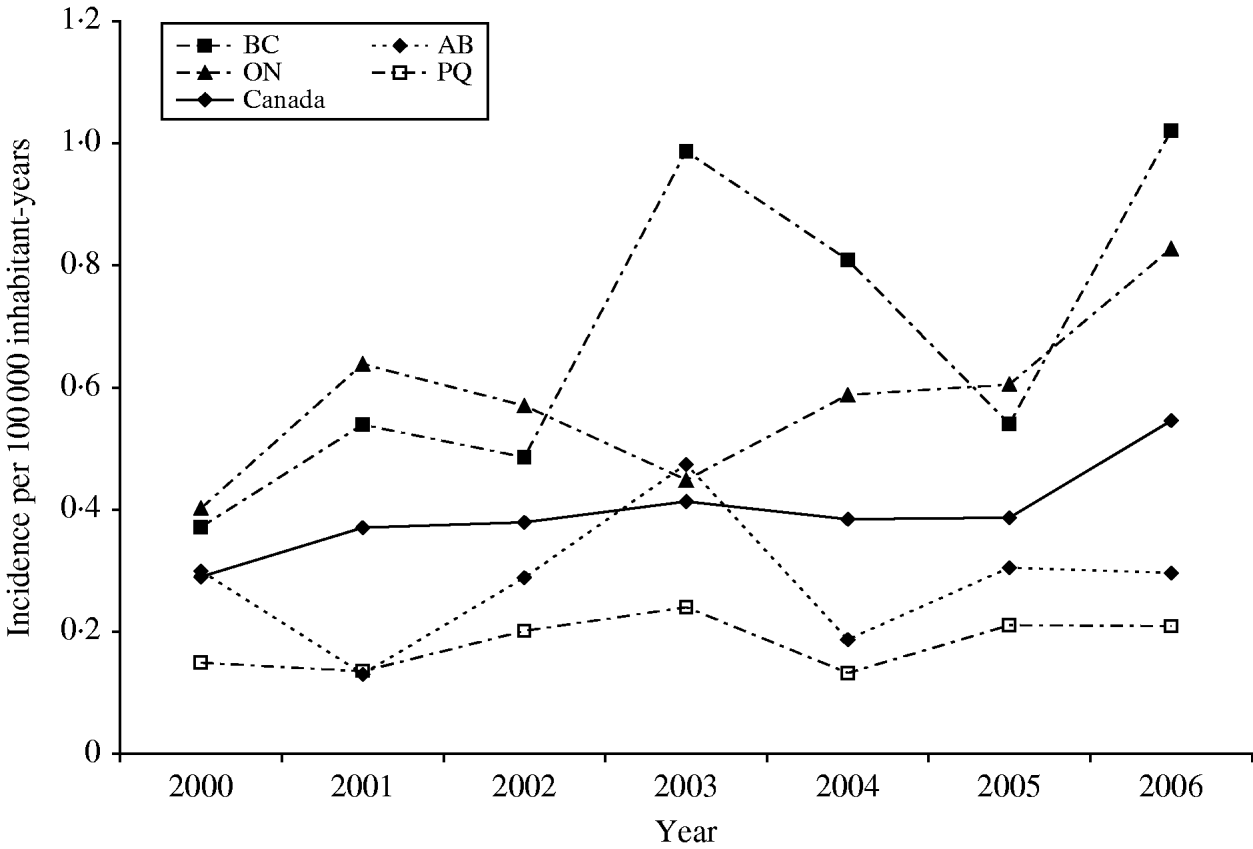
Fig. 1. Incidence rates of S. Typhi in Canada and in the four most populated provinces, 2000–2006. Province identification codes: BC, British Columbia; AB, Alberta; ON, Ontario; PQ, Québec.
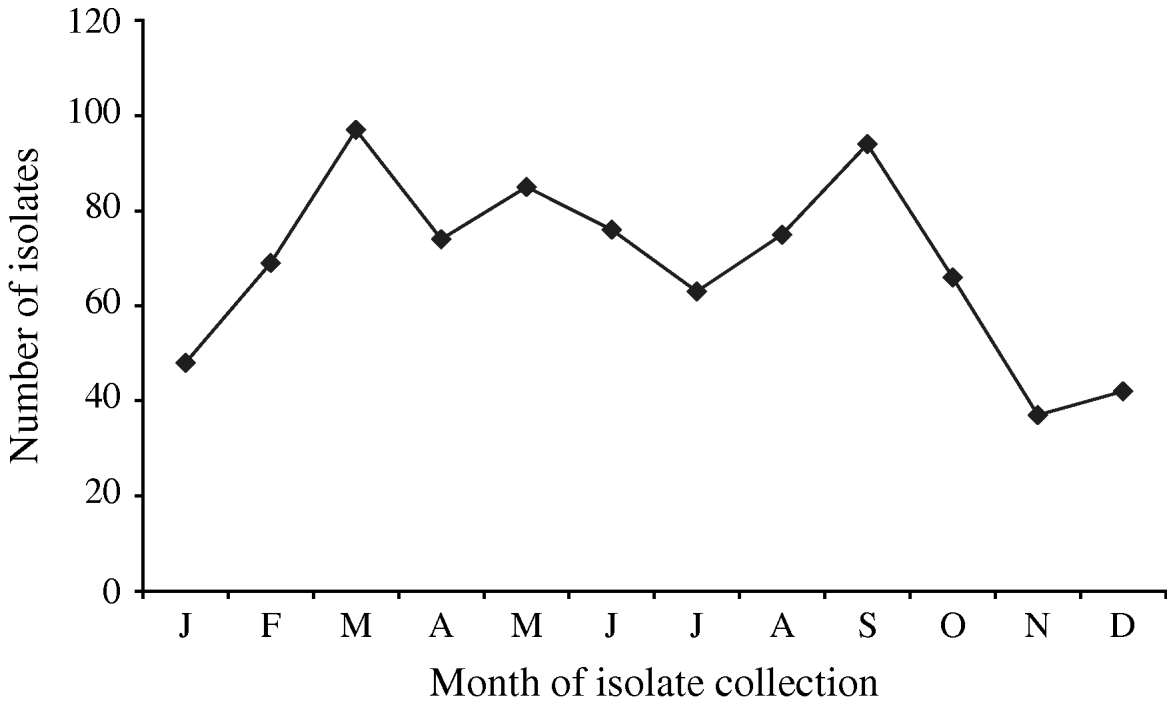
Fig. 2. Number of S. Typhi isolates submitted to the National Microbiology Laboratory in Canada by month of isolate collection, 2000–2006.
A total of 826 isolates were submitted to the NML for further analysis including phage typing, PFGE, and antimicrobial susceptibility testing. Of these, 432 (52%) isolates were from blood samples, 223 (27%) from stool, five (0·6%) from abscesses or wounds, four (0·5%) from urine, three (0·4%) from bodily fluids or aspirates, and 159 (19%) were from non-specified sources. Gender and age information were available for 78% (n=649) and 79% (n=655) of S. Typhi isolates, respectively. The median age was 23 years (range 0–98); 326 (50%) of cases were male. The highest isolation rates occurred in the 0–12 years age group with 0·59/100 000, and decreased with increasing age to 0·09/100 000 in the ⩾65 years age group (Fig. 3).
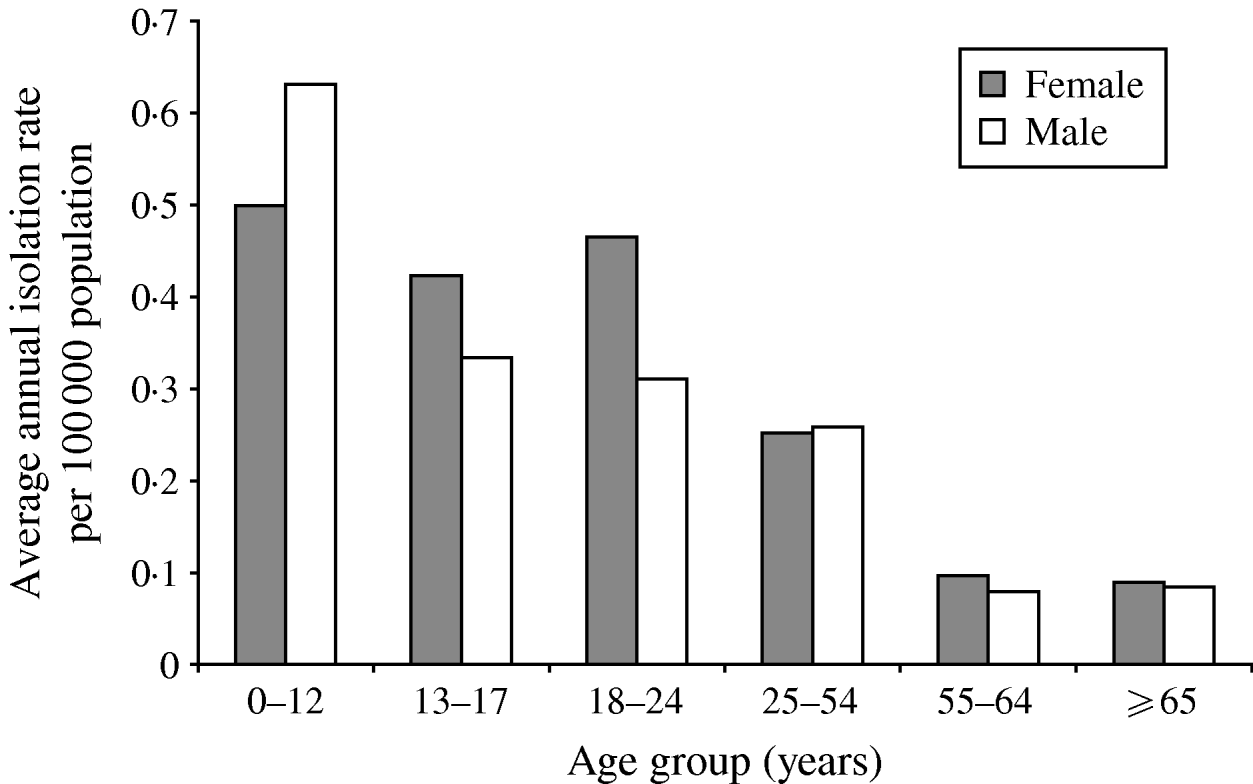
Fig. 3. Average annual age- and gender-specific isolation rates of S. Typhi in Canada, 2000–2006.
Antimicrobial resistance
Antimicrobial susceptibility testing was carried out on 826 S. Typhi isolates from 2000 to 2006. Resistance to one or more antimicrobials was detected in 61% (n=505) of the isolates tested, while resistance to three or more antimicrobial classes was observed in 148 (18%) of the isolates (Table 1). Overall, 56% (n=465) were resistant to NAL; 18% to AMP (n=150) or SUL (n=146); 17% to CHL (n=143), STR (n=144) or SXT (n=144); and 15% to TCY (n=126). Fewer than 1% were resistant to AMC (n=4), FOX (n=4), TIO (n=3) or KAN (n=2). No resistance was observed to AMK, CRO, CIP, and GEN (Table 1). Trends in the resistance to AMP, CHL, STR, TCY, and SXT were all highly similar to one another, fluctuating together throughout the study period and peaking in 2005 (Fig. 4).
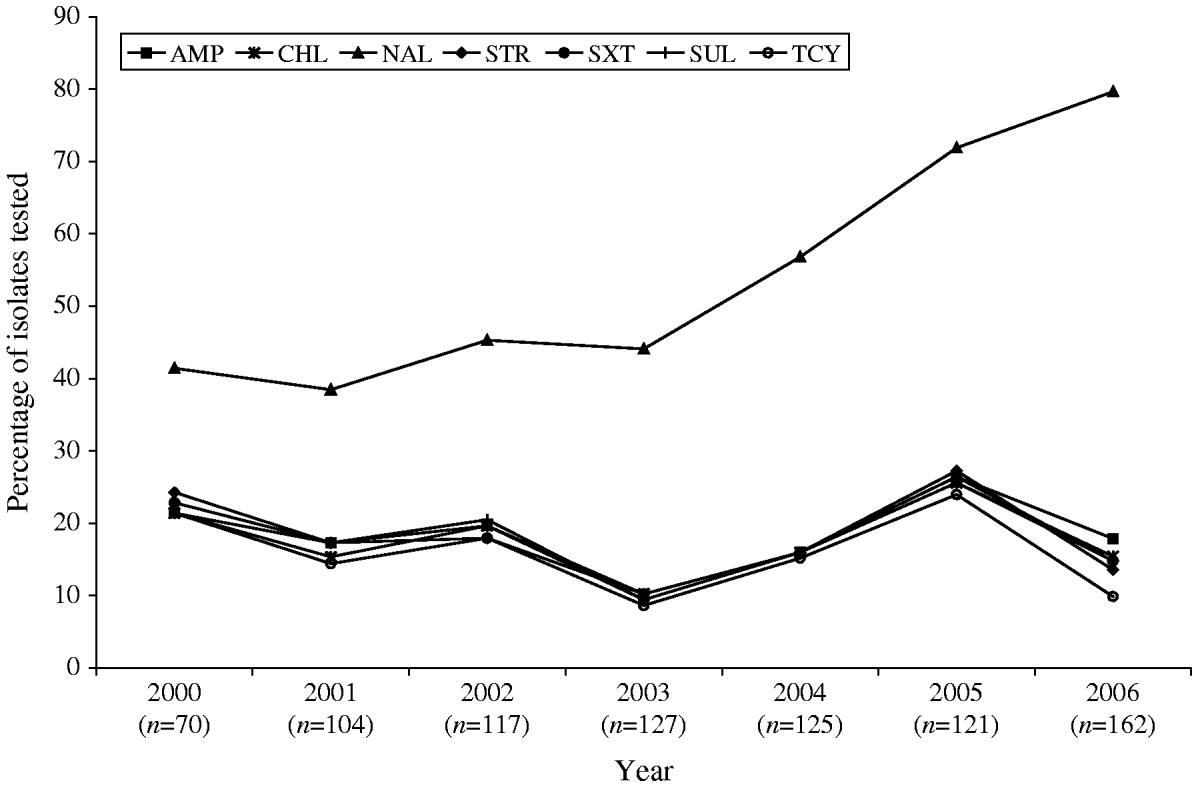
Fig. 4. Trends in resistance to selected antimicrobials in S. Typhi isolated in Canada, 2000–2006.
Table 1. Antimicrobial resistance patterns of S. Typhi isolated in Canada, 2000–2006
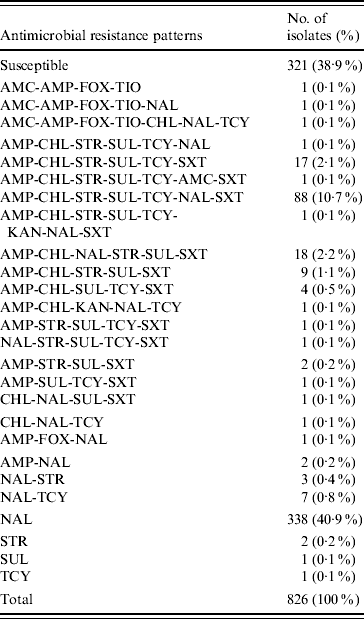
AMK, Amikacin; AMC, amoxicillin-clavulanic acid; AMP, ampicillin; FOX, cefoxitin; CRO, ceftriaxone; TIO, ceftiofur; CHL, chloramphenicol; CIP, ciprofloxacin; GEN, gentamicin; KAN, kanamycin; NAL, nalidixic acid; STR, streptomycin; SUL, sulfonamide; TCY, tetracycline.
A substantial increase in quinolone resistance in S. Typhi isolated in Canada was observed. During 2000–2006, the proportion of NAL-R S. Typhi strains increased significantly (P<0·001) from 41% (29/70) to 80% (129/162) (Fig. 4). Of 535 S. Typhi strains tested from 2003 to 2006, 343 (64%) were NAL-R, of which 337 (98%) had a minimum inhibitory concentration (MIC) value for CIP of ⩾0·125 μg/ml, demonstrating a relationship between NAL-R and reduced susceptibility to CIP (P<0·0001) (Fig. 5).
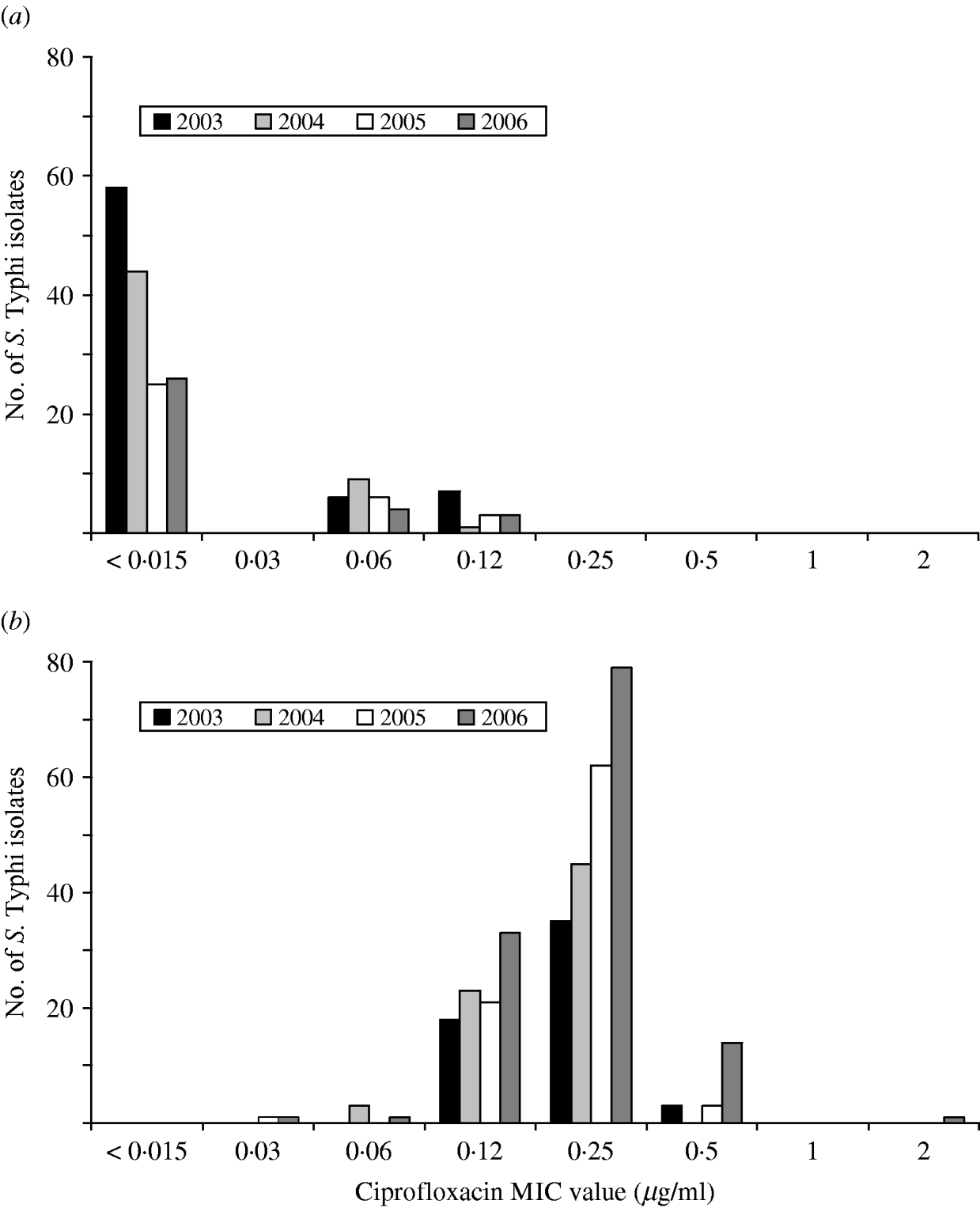
Fig. 5. Ciprofloxacin MIC values for (a) nalidixic acid-susceptible and (b) nalidixic acid-resistant isolates of S. Typhi in Canada, 2003–2006.
Of the 26 resistance profiles observed the most prevalent was resistance to NAL alone which accounted for 338 isolates (41%), followed by 88 (11%) isolates with the AMP-CHL-STR-SUL-TCY-NAL-SXT resistance profile, and 18 (2%) isolates of the AMP-CHL-NAL-STR-SUL-SXT resistance profile. The AMP-CHL-SXT phenotype was observed in 138 (17%) of the S. Typhi isolates, and AMP-CHL-STR-SUL-TCY was recorded in 108 (13%), always in combination with resistance to other antimicrobials (Table 1). The proportion of MDR S. Typhi strains in Canada has fluctuated from 23% (16/70) of the strains in 2000, declining to a low in 2004 of 10% (13/127), and then increasing again to 26% (32/121) in 2006.
Phage typing
Thirty-nine phage types were identified in the 826 S. Typhi isolates analysed during 2000–2006. There were 134 phage type (PT) Untypable strains that included 65 Untypable Vi Negative, 45 Untypable Vi Positive and 24 Degraded Vi Positive strains (Table 2). The most common phage type identified was PT E1 comprising 39% (n=324) of the isolates tested (Table 2). PT E9 ranked a distant second, accounting for 7% (n=54) followed by PT A (6%, n=51), PT E14 (4%, n=35) and PT O (4%, n=29). PT M1, an epidemic phage type in South and Central Asia [Reference Rowe, Ward and Threllfall9, Reference Hampton12], ranked 11th representing only 2% (n=15) of the S. Typhi strains tested. The proportion of isolates identified as PT E1 decreased from 49% (34/70) of isolates in 2000 to 39% (63/162) in 2006 (Fig. 6). PT E9 increased from 4% (3/70) in 2000 to 14% (17/125) in 2004, and then decreased to 5% (8/162) in 2006. PT A also decreased from 7% (5/70) in 2000 to 5% (8/162) in 2006 (Fig. 6).
Molecular characterization (PFGE)
Analysis of 222 isolates by PFGE using enzyme XbaI resulted in 91 distinguishable patterns, demonstrating a moderate level of genetic diversity (Table 2). The five most frequently occurring PFGE patterns accounted for 46% (n=102) of all isolates indicating better discriminatory power than phage typing, wherein the five most frequently occurring phage types identified during 2000 to 2006 comprised 60% (n=493) of all isolates (Table 2). The PFGE patterns had an overall similarity of 66% and clustered into four main groups (Fig. 7). The majority of isolates (67%, n=148) clustered into group 2, the patterns of which showed the highest level of overall similarity (84·79%) compared to the other groups (group 1A, 82·92%; group 1B, 82·19%; group 3, 68·4%). Nearly all isolates in group 3 had unique PFGE patterns (Fig. 7).
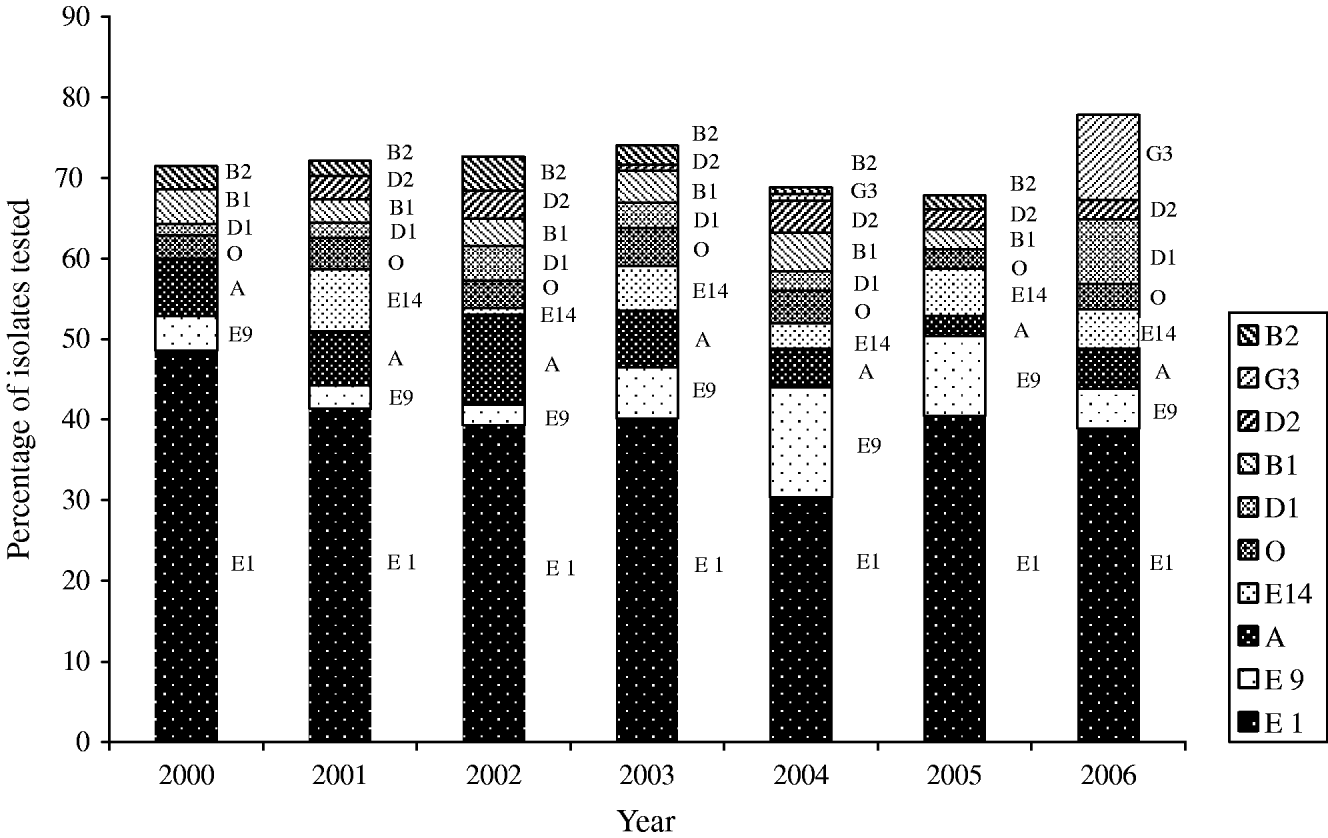
Fig. 6. Trends of the frequently isolated phage types of S. Typhi in Canada, 2000–2006.
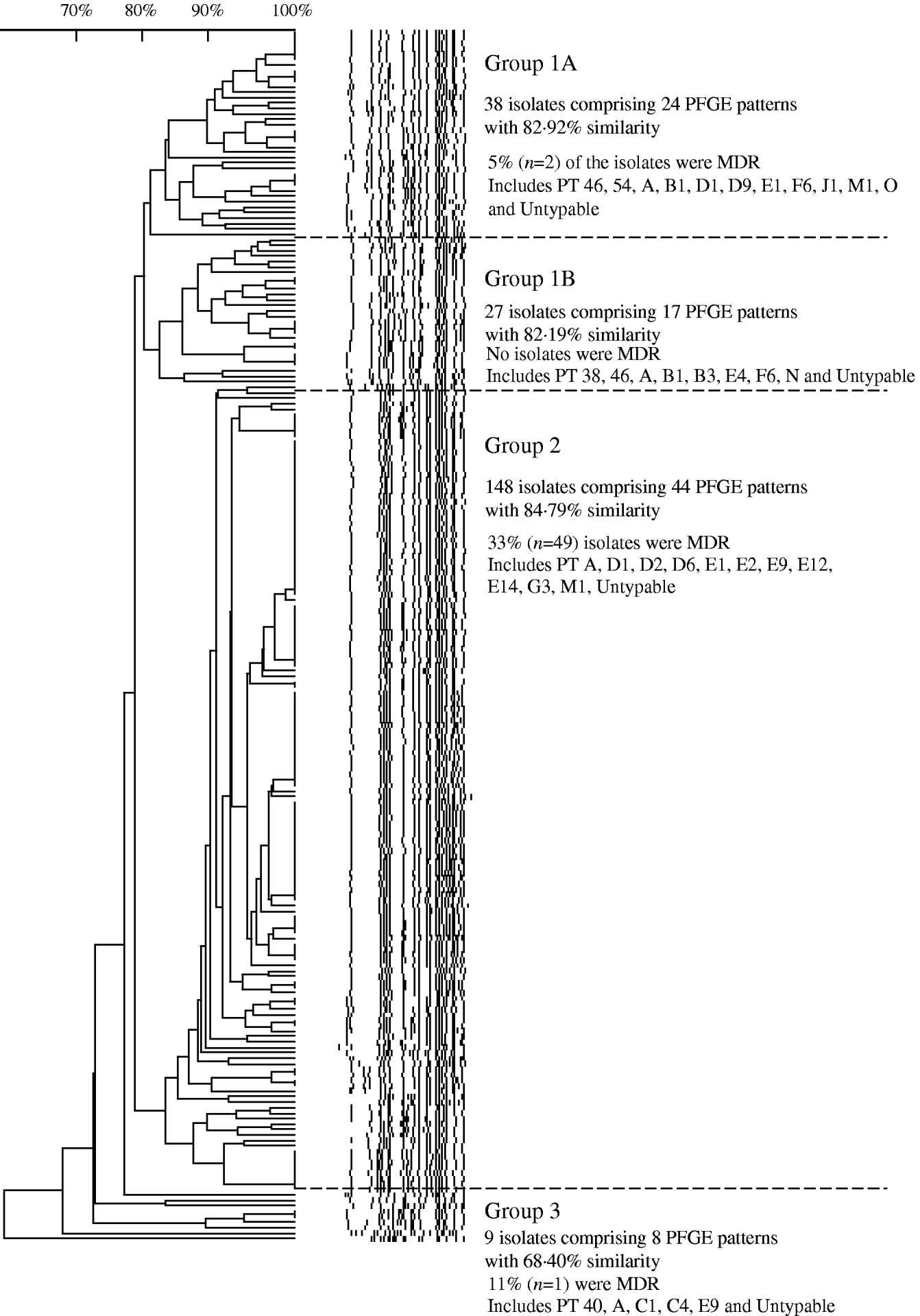
Fig. 7. Similarity of S. Typhi PFGE patterns isolated in Canada, 2004–2005.
Table 2. Diversity of PFGE patterns and phage types of S. Typhi isolated in Canada
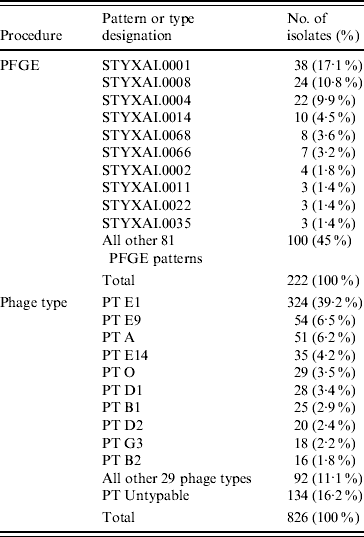
Table 3. Distribution of S. Typhi phage types in the major PFGE groups, 2004–2005
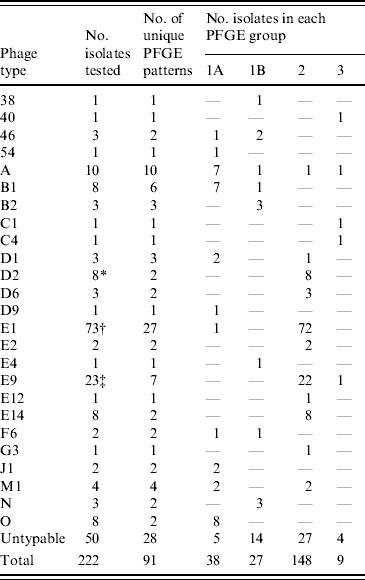
* Seven (88%) isolates had the same PFGE pattern.
† Forty-five (62%) isolates had one of two patterns.
‡ Seventeen (74%) isolates had the same pattern.
Table 4. Antimicrobial resistance profiles correlated to the S. Typhi phage types identified during 2000–2006
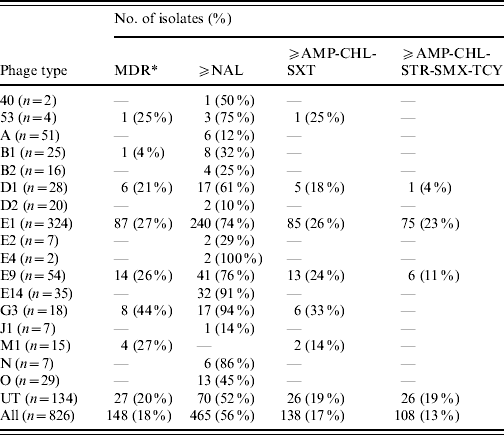
MDR, Multidrug resistant is defined as resistance to three or more classes of antibiotics based on mechanism of action, that include: aminoglycoside (AMK, GEN, KAN, STR); β-lactamase (AMC, AMP, FOX, CRO, TIO); chloramphenicol (CHL); quinolone (NAL, CIP); sulfonamide (SMX, SSS, SUL); tetracycline (TCY); trimethoprim–sulfamethoxazole (SXT).
Foreign-acquired infections
Travel history was available for 63 cases: 41 (65%) of these cases reported travel to India; 11 (17%) travel to Pakistan; three (5%) to Bangladesh; two each to the Philippines and Haiti; and one case from each for Cambodia, Guatemala, South Africa and Mexico. The average annual incidence of notified infections due to S. Typhi over the study period was highest in Canadian residents traveling to Pakistan (2·90 cases/100 000 Canadian resident travellers), followed by travel to Bangladesh (1·88), Haiti (1·42), and India (1·25). Lower rates were observed for the Philippines (0·08), Cambodia (0·15), Guatemala (0·18), Mexico (0·002) and South Africa (0·05).
Twenty-six (41%) of the travel-related isolates were PT E1, 18 of which were associated with travel to India and five to Pakistan. One PT E1 isolate was associated with travel to each of Cambodia, Guatemala and Mexico. All five (8%) of the travel-related PT E9 isolates reported India as a travel destination. Seventeen isolates (27%) were distributed in 10 other phage types and 15 (24%) were PT Untypable.
Ten MDR isolates were associated with travel to India (n=4), Pakistan (n=4), Bangladesh (n=1), or Cambodia (n=1). Nine of these isolates had the AMP-CHL-STR-SUL-TCY resistance profile with additional resistances to NAL-SXT (8/9) and SXT (1/9). Forty-seven (75%) isolates were resistant to NAL, alone (36/47) or in addition to other antimicrobials (3/47). Of these, 37 (79%) travelled to India, six (13%) to Pakistan, three (6%) to Bangladesh, and one (2%) to Cambodia. Isolates from patients with a travel history to the Indian subcontinent accounted for 9/10 MDR and 46/47 NAL-R travel-related S. Typhi isolates. Furthermore, the presence of NAL-R was 11 times greater (P<0·001, 95% CI 2·9–42·0) in isolates of cases that travelled to India than in isolates from cases that travelled to other areas of the world.
PFGE patterns were available for 19 of the cases with travel history, among those there were 13 unique PFGE patterns. Ten isolates with travel history to India had 10 different PFGE patterns; however, two PFGE patterns from India were also isolated from cases reporting travel to Pakistan (STYXAI.0001, STYXAI.0004). PFGE group 2 accounted for most of the travel-related isolates including India (n=7), Pakistan (n=6) and the Philippines (n=1). There were two isolates of PFGE similarity group 1B from each of Haiti and India, as well as one PFGE group 1A isolate from India.
DISCUSSION
Although the level of typhoid infection, and subsequently the burden of illness, in Canada are low, foreign-acquired infections continue to be of concern by placing additional demands for resources upon the public healthcare system. Treating S. Typhi infections caused by resistant strains places a larger burden on public health due to the requirement for more extensive therapies, which tend to be less effective and often result in longer stool carriage rates than treatments for susceptible strains [Reference Connor and Schwartz4]. Of particular concern is the acquisition of multiple drug resistance and decreased susceptibility to quinolone agents, as these are the drugs of choice for treating severe or acute salmonellosis. Although full resistance to ciprofloxacin was not observed in this study, a steady increase in NAL-R was coupled to a reduction in ciprofloxacin susceptibility over the 7-year period. In 2006, 80% of the S. Typhi isolates were NAL-R, 98% of which had reduced susceptibility to ciprofloxacin. The presence of typhoid isolates with resistance to ciprofloxacin has been reported in 4% of S. Typhi tested in Nepal between 2002 and 2004 [Reference Chau14]. In the USA, 42% of the S. Typhi isolates in 2004 were NAL-R, a large increase from 19% observed in 1999; and reduced susceptibility to ciprofloxacin was present in 42% of isolates in 2004 [41]. Similarly, in the UK, 32% of isolates were NAL-R and had reduced susceptibility to ciprofloxacin during 2000–2003 [Reference Cooke23]. In 2006, 26% of S. Typhi isolates in Canada were MDR representing a relatively lower risk to public health in contrast with endemic regions where increasing prevalence is observed. In Asia the proportion of S. Typhi with MDR has increased from 63% in 1992 to >90% by early 2000 [Reference Chau14].
While PFGE is the current, internationally standardized method of molecular subtyping for S. Typhi [Reference Swaminathan42], phage typing has had a long history as a subtyping technique for global surveillance [Reference Anderson and Williams21] and has substantial historical value; thus both the phenotypic and genotypic subtyping methods are routinely applied. Analysis of the relationship of S. Typhi PFGE and phage types indicated that the majority of isolates from the same phage type generally grouped into common PFGE similarity groups (1A, 1B, 2 or 3), although this was not absolute (Table 3). A major advantage of the application of PFGE subtyping is that it enables the characterization of isolates that are not typable by phage typing. PFGE analysis of 50 PT Untypable strains resulted in 28 distinct DNA patterns, generally grouped evenly into all the PFGE relatedness groups. Half of the PT Untypable isolates had unique DNA patterns, whereas some matched patterns were strongly associated with a particular phage type, such as phage types A, B1, D1, E1, and F6. Owing to its increased discriminatory power, PFGE subtyping also facilitated better epidemiological linkages between clinical cases.
Slightly lower diversity of PFGE patterns was observed in drug-resistant isolates (72% similarity) compared to drug-susceptible isolates (68% similarity), indicating that resistant isolates were genetically highly related, particularly those with the MDR phenotype. The PFGE similarity group 2 (corresponding to phage types D and E Vi) had the highest level of MDR isolates (33%) and a higher proportion of NAL-R strains (75%) than did any of the other groups. This is consistent with previous findings regarding the clonal nature of MDR S. Typhi [Reference Kubota5, Reference Thong, Bhutta and Pang20, Reference Fica43]. Further analysis of PFGE patterns of MDR S. Typhi isolated in Canada may help to further elucidate their evolution; recent work suggests that the MDR phenotype emerged relatively recently due to the clonal expansion of a small number of strains that may have acquired plasmid-mediated resistance genes [Reference Kubota5, Reference Mirza44].
Similarly, the distribution of AMR profiles in phage types was also non-homogeneous. Overall, the majority of NAL-R and MDR isolates were PT E1, which is the most prevalent phage type in Canada, accounting for 39% of strains tested. However, when stratified by phage type, PT G3 and PT E14 had the highest proportion of NAL-R isolates (Table 4).
In Canada, travel information is requested to accompany isolates submitted to the national surveillance systems; however, in practice, little of this type of information is relayed through the reporting chain from the local laboratories to federal level. Information collected through this study has shown PT E1 to be the most predominant phage type, accounting for 41% of the travel-related isolates, the majority of which were associated with travel to India and Pakistan. This phage type is also predominant in regions of the world where typhoid is endemic and is the most commonly encountered phage type of S. Typhi isolated from travel-associated cases in the UK [Reference Cooke23] and the USA [Reference Mermin45].
Conclusions regarding any geographic clustering of PFGE patterns were limited by the amount of travel information available. Nevertheless, our results showed a diversity of strains occurring within a single country (India) plus some PFGE patterns that were associated with more than one country, corroborating previous studies [Reference Thong46]. In their analysis of travel-associated S. Typhi PFGE patterns, Kubota et al. [Reference Kubota5] found significant strain diversity within single countries, particularly within the Indian subcontinent as well as PFGE types that were associated with geographically diverse locales. This group also found a predominant strain of MDR S. Typhi circulating in India, Pakistan, and Vietnam, isolates of which were indistinguishable by their PFGE pattern. Since standardized PulseNet methodology is used in Canada, the USA, and increasingly in other countries via the PulseNet International network, direct comparisons of patterns is possible. The creation of a global S. Typhi database will facilitate comparisons of isolates from travel-associated cases in Canada with patterns isolated elsewhere.
In Canada, typhoid infections are assumed to be foreign-acquired and are therefore of concern. In the USA, international travel has been recognized as the most significant risk factor for S. Typhi infections since the 1970s [Reference Crump15, Reference Mermin45, Reference Rice, Baine and Gangarosa47, Reference Ryan, Hargrett-Bean and Blake48]. Further, between 2000 and 2003, 60% of all S. Typhi isolates in England, Scotland, and Wales were known to have been travel-associated, the vast majority of which (70%) reported India or Pakistan as the place of acquisition of infection [Reference Cooke23]. Other estimates indicate that about 80% of cases in the USA are acquired outside the country, primarily from the Indian subcontinent and south-east Asia [Reference Navarro25]. Country of ethnicity may contribute to the overall incidence of typhoid cases by influencing the extent of cultural immersion in these countries, a difference which has been observed between tourists and those visiting friends and relatives [Reference Crump, Luby and Mintz1, 49–Reference Ochiai51]. Studies on this latter group indicated that they do not perceive that they are at risk of developing typhoid fever, making them more likely to drink untreated water and eat undercooked foods, and more likely to travel without visiting a travel clinic or seeking medical advice on appropriate vaccinations [49, Reference Angell and Cetron52]. A review of travel-related typhoid fever cases showed that 4% of cases were tourists, while 40% were travellers that had visited friends and relatives [Reference Ackers11]. Of these, only 4% had received vaccinations before travel. Similar trends have been observed in Canada, where only a small proportion, <15% of travellers, visit travel clinics before departure [Reference Keystone53].
CONCLUSIONS
Although typhoid fever is not endemic to Canada, it represents a challenge to public health due to increasing travel and globalization. Surveillance of S. Typhi infections enables us to establish baseline incident rates, characterize the strains encountered within the Canadian population, and gain insight on the characteristics of S. Typhi acquired by Canadians via travel. The ability to track shifts in genotypic and phenotypic strain occurrence in real time can be used to predict global transmission patterns. Due to its potential impact upon public health, it is of particular importance to be able to recognize unusual or emerging strains that may harbour potentially significant virulence characteristics (e.g. antimicrobial resistance). Of particular concern is the acquisition of multiple drug resistance and decreased susceptibility to fluoroquinolone agents, as these are the drugs of choice for treating severe or acute salmonellosis.
Further efforts are required to encourage the dissemination of patient travel information through the surveillance system to better distinguish locally acquired from foreign-acquired infections. Enhanced epidemiological information and laboratory characterization of foreign-acquired infections can provide enhanced risk estimates associated with travel to specific regions of the globe. These risk estimates may then be used to inform and educate travellers in an attempt to reduce the number of people returning to Canada with infections acquired abroad, and thereby reducing healthcare costs incurred by the domestic treatment of these infections. These risk estimates may also be used to inform clinicians when considering treatment options.
Due to the high global burden of typhoid infections and the increasing development and presence of antimicrobial resistance, the World Health Organization has recently updated their position on typhoid vaccines. Countries with endemic typhoid fever are encouraged to consider creating mitigation programmes and provide typhoid vaccinations to control endemic disease as well as outbreaks [Reference Guzman6, 54]. Similarly, travellers should be offered and consider vaccinations, particularly those visiting areas where the risk of typhoid infection is high and where AMR strains are prevalent.
ACKNOWLEDGEMENTS
The authors acknowledge provincial epidemiologists from the provinces of Ontario, British Columbia, Quebec, and Alberta for providing additional information, Rachel McCormick for reading and providing input into the paper, Cynthia Misfeldt, Helen Tabor, Rafiq Ahmed, Derek Ozunko, Geoff Soule, Gail Christie, Devon Kuntz, Michael Mulvey, and Denis Le for performing PFGE, phage typing, serotyping, and antimicrobial resistance testing at the National Microbiology Laboratory. We also acknowledge the contribution provided by all the Provincial Public Health Laboratories for submitting isolates for further characterization and antimicrobial susceptibility testing. The Canadian Integrated Program for Antimicrobial Resistance Surveillance Public Health Partnership and the Canadian Public Health Laboratory Network include the provincial public health laboratories or enteric reference centres of British Columbia, Alberta, Saskatchewan, Manitoba, Ontario, Québec, New Brunswick, Nova Scotia, Prince Edward Island and Newfoundland and Labrador; the Laboratory for Food-borne Zoonoses, Guelph, Ontario; the National Microbiology Laboratory, Winnipeg, Manitoba; and the Center for Food-borne, Environmental and Zoonotic Infectious Diseases, Guelph, Ontario.
DECLARATION OF INTEREST
None.













