To the Editor—Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can result in prolonged polymerase chain reaction (PCR) positivity, particularly in the immunocompromised population. Reference Wang, Xu and Zhang1–Reference Helleberg, Niemann and Moestrup4 This prolonged positivity poses infection prevention and public health management issues because the infectivity and transmission risks are unclear.
The state of Victoria, Australia (population 6.4 million), experienced a second wave of coronavirus disease 2019 (COVID-19) from June to October 2020. 5 To combat it, public health measures were implemented that included (at most restrictive stage) mandatory mask use, limitations in travel to 5 km, no household visitors, limited outdoor activities, and night-time curfews. Here, we report a case with late symptom onset of COVID-19 in an immunocompromised patient and the public health implications of prolonged isolation in a low-prevalence community setting.
A 53-year-old man with follicular non-Hodgkin lymphoma receiving a CD3-CD20 bispecific antibody (bsAb) therapy on clinical trial was tested for SARS-CoV-2 in the setting of close contact with his son, who was diagnosed with COVID-19 on July 24, 2020. On July 28, the patient’s asymptomatic screening swab (combined deep nasal/oropharyngeal) tested on a respiratory multiplex panel (AusDiagnostics Respiratory Pathogens 16-well panel, Mascot, Australia) detected SARS-CoV-2.
The patient was directed to isolate at home, and bsAb therapy was halted. Contact tracing identified 26 community and healthcare close contacts who were quarantined, but no transmission of SARS-CoV-2 was identified.
Over the next 2 months, the patient remained asymptomatic in the community and had regular PCR testing. State and national clearance guidelines at the time required 2 negative SARS-CoV-2 PCR tests in those with significant immunocompromise. 6,7
Virus was detectable on PCR at each time point, and Table 1 shows the cycle threshold (Ct) values for each episode of testing (timeline also represented in Fig. 1).
Table 1. Microbiologic Testing Results
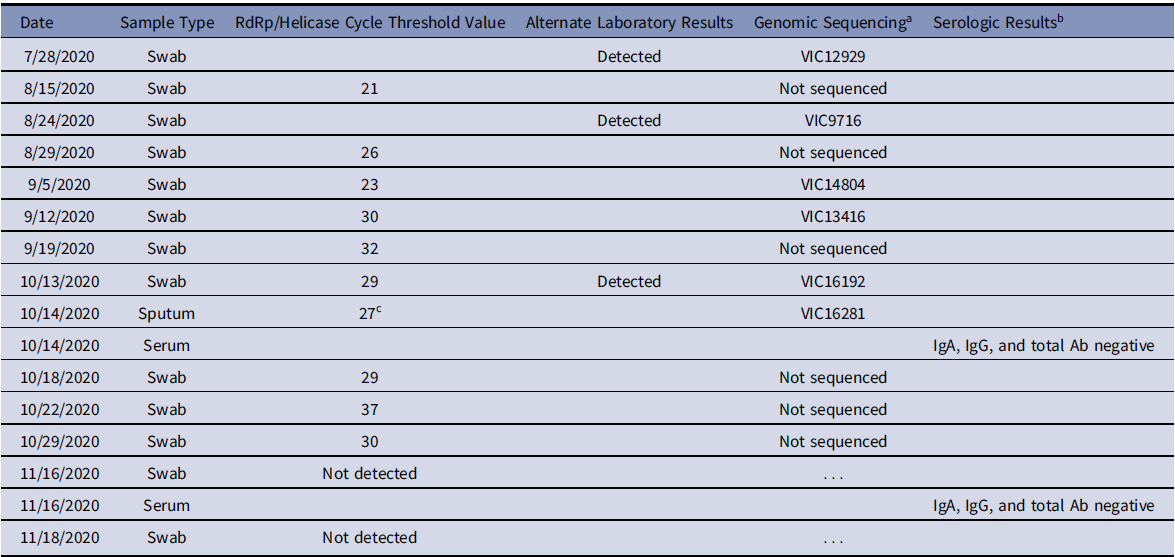
a Sequence identifier in GISAID (https://www.gisaid.org/) and NCBI SRA (https://www.ncbi.nlm.nih.gov/sra).
b Serologic assays Euroimmun SARS-CoV-2 S1 IgA and IgG EIA (Euroimmun, Lübeck, Germany) and Wantai SARS-CoV-2 Total Ab EIA (Wantai Biological Pharmacy, Beijing, China).
c Viral culture successful on this sample.

Figure 1. Summary of patient’s clinical course. PCR: polymerase chain reaction; D: day; Cd3-Cd20 Ab: CD3-CD20 T-cell engager bispecific antibody; Nø: neutrophil; Pt: patient; Dx: diagnosed; +ve: positive; −ve: negative; Ct cycle threshold; IV: intravenous.
On September 20, an expert panel of physicians decided that, given the length of time since initial diagnosis (54 days), lack of symptoms throughout, and increasing Ct values on PCR, infectivity was likely to be low. The patient was given an exemption to leave his house for short periods, which included resuming clinical trial therapy with hospital staff using droplet respiratory precautions. The bsAb was recommenced during overnight admission on October 6. Given previous hypersensitivity reaction to B-cell depleting therapies and long dose delay, premedication including 20 mg IV dexamethasone was administered with ramp-up schedule of bsAb (commencing 3% of usual dose).
One day after discharge (72 days after original COVID-19 diagnosis), the patient developed a new productive cough associated with dyspnea on exertion but no other symptoms. He was tachycardic (108 beats per minute) and tachypnoiec (22 breaths per minute), but other observations were within normal limits. Chest auscultation revealed fine crepitations at the right base; otherwise, examination was unremarkable. A chest computed tomography (CT) scan showed bilateral extensive peribronchial ground glass attenuation most consistent with atypical or viral infection. He was admitted and managed under droplet, contact, and airborne precautions.
Nasopharyngeal/oropharyngeal swab and expectorated sputum sample tested on the AusDiagnostics respiratory multiplex PCR panel were positive for SARS-CoV-2 (Table 1), with all other targets not detected. Bacterial sputum microscopy and culture yielded no pathogens. Virus isolation was attempted from sputum and nasopharyngeal/oropharyngeal swabs in Vero cells. SARS-CoV-2 was isolated from the sputum sample. The expert panel was reconvened on October 14, and they determined that the patient should be managed as an active, potentially infectious case of COVID-19, linked to the initial diagnosis from July 28.
SARS-CoV-2 genomic sequencing was performed on original patient samples by tiled amplicon PCR and Illumina sequencing, as previously described. Reference Seemann, Lane and Sherry8 The 4 samples prior to symptom onset and 2 samples after symptom onset were closely related by genomics with some evidence of minor in-host evolution, consistent with prolonged infection rather than alternative lineage reinfection.
Serology tests were performed on October 14 and November 16. SARS-CoV-2 IgA, IgG, and total antibody were not detected (Table 1). The patient was hypogammaglobulinemic throughout.
While in the hospital, the patient received 3 units of ABO-compatible convalescent plasma through compassionate access on October 16 and 17. He did not receive remdesivir, dexamethasone, or antibiotics. Cough and shortness of breath resolved, and he was discharged home on October 18. The patient was required to isolate at home, and subsequent swabs detected SARS-CoV-2 on PCR until November 16, when his test result was “not detected” for the first time (Table 1). Swabs following discharge have not demonstrated culturable virus.
This is an unusual case of SARS-CoV-2 infection with late symptom onset (72 days after initial diagnosis) in an immunocompromised patient. Possible triggers for this include bsAb therapy, cytokine release syndrome, or dexamethasone use. The former 2 factors are unlikely given the very low dose of bsAb and the lower Ct values for SARS-CoV-2 PCR and culturable virus.
The virus was still culturable, which raised concerns about transmissibility. The patient continued (until November 16, 111 days) to have prolonged PCR positivity with no development of antibodies to SARS-CoV-2 and did not meet hospital or statewide guidelines for clearance.
Given the transition to elimination of sustained community transmission of COVID-19 in Victoria, a very cautious approach was taken to the isolation and public health management of this case. The fact that there was no evidence that this case infected any close household or other contacts over an extended period raises the question of whether prolonged infection, even with culturable virus, poses an ongoing infection risk to others, and whether prolonged isolation with potential significant negative psychological effects is justified. Reference Brooks, Webster and Smith9 More research into this important issue is needed.
Acknowledgments
We thank the patient and his family for their consent and participation in this report. We thank James Daly (Australian Red Cross Lifeblood) for his assistance with the provision of convalescent plasma for the patient.
Financial support
No financial support was provided relevant to this article.
Conflicts of interest
G.P.G. is an advisory board member for Novartis, Roche, Gilead, Sandoz, and Janssen and has received research funding from Janssen, BeiGene, AstraZeneca, AbbVie, and MSD.





