Introduction
Kinetoplastids are one of three major groups of organisms that belong to the evolutionarily divergent protist phylum Euglenozoa (Cavalier-Smith, Reference Cavalier-Smith2016). Although divergent, euglenozoans are ubiquitous; representatives from all three groups are easily isolated from many freshwater, marine and soil environments and, in part due to their overall abundance, contribute significantly to ecosystem ecology (von der Heyden et al., Reference von der Heyden, Chao, Vickerman and Cavalier-Smith2004; Edgcomb et al., Reference Edgcomb, Orsi, Breiner, Stock, Filker, Yakimov and Stoeck2011; Lukeš et al., Reference Lukeš, Flegontova and Horák2015; Mukherjee et al., Reference Mukherjee, Hodoki and Nakano2015; Flegontova et al., Reference Flegontova, Flegontov, Malviya, Audic, Wincker, de Vargas, Bowler, Lukeš and Horák2016; Flegontova et al., Reference Flegontova, Flegontov, Malviya, Poulain, de Vargas, Bowler, Lukeš and Horák2018).
Systematically, the kinetoplastids separate into the monophyletic, obligatory parasitic trypanosomatids and a wide diversity of free-living, bi-flagellate phagotrophs, with occasional examples of parasites and symbionts populating three major clades (von der Heyden et al., Reference von der Heyden, Chao, Vickerman and Cavalier-Smith2004; Simpson et al., Reference Simpson, Stevens and Lukeš2006; Kaufer et al., Reference Kaufer, Ellis, Stark and Barratt2017; Yazaki et al., Reference Yazaki, Ishikawa, Kume, Kumagai, Kamaishi, Tanifuji, Hashimoto and Inagaki2017). It is the uniflagellate trypanosomatids that are the best known due to the role of some as the aetiological agents of serious, neglected tropical diseases (Nussbaum et al., Reference Nussbaum, Honek, Cadmus and Efferth2010). The defining characteristic common to both free-living and parasitic kinetoplastids is the coalescence (in trypanosomatids the catenation) of several thousand circular DNA molecules to form distinctive mitochondrial genome architectures, known more commonly as kinetoplasts, and which give rise to the class name Kinetoplastea (Lukeš et al., Reference Lukeš, Archibald, Keeling, Doolittle and Gray2002). Uridine-insertion and -deletion editing of mRNA on a massive scale is essential for gene expression from these genomes and provides a second example of extreme or unusual biology that defines and pervades throughout the kinetoplastids (Aphasizhev and Aphasizheva, Reference Aphasizhev and Aphasizheva2014; David et al., Reference David, Flegontov, Gerasimov, Tanifuji, Hashimi, Logacheva, Maruyama, Onodera, Gray, Archibald and Lukeš2015; Read et al., Reference Read, Lukeš and Hashimi2016). For further examples of extreme kinetoplastid biology that have peripheral relevance for this review – peroxisome-compartmentalized carbohydrate metabolism, loss of transcriptional control on protein-coding gene expression, flagellar pocket dynamics – readers are directed towards articles by Haanstra et al. (Reference Haanstra, González-Marcano, Gualdrón-López and Michels2016), Morales et al. (Reference Morales, Hashimoto, Williams, Hirawake-Mogi, Makiuchi, Tsubouchi, Kaga, Taka, Fujimura, Koike, Mita, Bringaud, Concepción, Hashimoto, Embley and Nara2016a), Clayton (Reference Clayton2014), and Field and Carrington (Reference Field and Carrington2009).
Although trypanosomatid species are widely known as the causative agents for diseases of medical, veterinary and agricultural importance (Jaskowska et al., Reference Jaskowska, Butler, Preston and Kelly2015; Giordani et al., Reference Giordani, Morrison, Rowan, de Koning and Barrett2016; Field et al., Reference Field, Horn, Fairlamb, Ferguson, Gray, Read, De Rycker, Torrie, Wyatt, Wyllie and Gilbert2017; Kaufer et al., Reference Kaufer, Ellis, Stark and Barratt2017), most members of the family are simply monoxenous parasites of insects (Podlipaev et al., Reference Podlipaev, Sturm, Fiala, Fernandes, Westenberger, Dollet, Campbell and Lukes2004; Maslov et al., Reference Maslov, Votýpka, Yurchenko and Lukeš2013; Lukeš et al., Reference Lukeš, Skalický, Týč, Votýpka and Yurchenko2014; Kaufer et al., Reference Kaufer, Ellis, Stark and Barratt2017) with not always a clear indication that these protists are pathogenic towards their invertebrate host(s). Also less widely recognized is that at least twice, the symbiosis between a bacterial endosymbiont and a host trypanosomatid has occurred (Du et al., Reference Du, Maslov and Chang1994; de Souza and Motta, Reference de Souza and Motta1999; Votýpka et al., Reference Votýpka, Kostygov, Kraeva, Grybchuk-Ieremenko, Tesařová, Grybchuk, Lukeš and Yurchenko2014; Kostygov et al., Reference Kostygov, Butenko, Nenarokova, Tashyreva, Flegontov, Lukeš and Yurchenko2016) (Fig. 1). Trypanosomatid taxa involved in these events are not particularly closely related, and different evolutionary trajectories are possibly evident for each symbiosis: in the Strigomonadinae, growth and division of a single bacterial endosymbiont is entrained within the cell cycle of the host cell (Motta et al., Reference Motta, Martins, de Souza, Catta-Preta, Silva, Klein, de Almeida, de Lima Cunha, Ciapina, Brocchi, Colabardini, de Araujo Lima, Machado, de Almeida Soares, Probst, de Menezes, Thompson, Bartholomeu, Gradia, Pavoni, Grisard, Fantinatti-Garboggini, Marchini, Rodrigues-Luiz, Wagner, Goldman, Fietto, Elias, Goldman, Sagot, Pereira, Stoco, de Mendonça-Neto, Teixeira, Maciel, de Oliveira Mendes, Ürményi, de Souza, Schenkman and de Vasconcelos2010), whereas in recently discovered Novymonas less stringent regulation on the number of β-proteobacterial Pandoraea endosymbionts could reflect either symbiont farming or a snap-shot of an early transitional phase in the establishment of a novel endosymbiont–host relationship (Kostygov et al., Reference Kostygov, Butenko, Nenarokova, Tashyreva, Flegontov, Lukeš and Yurchenko2016, Reference Kostygov, Dobáková, Grybchuk-Ieremenko, Váhala, Maslov, Votýpka, Lukeš and Yurchenko2017). In this mini-review, we consider metabolic advantages conferred by bacterial endosymbionts to their partner trypanosomatids, how the biology of the host cell potentially influences the establishment, reductive evolution and subsequent entrainment of the endosymbiont(s), and we survey the literature with regard to endosymbioses within free-living phagotrophic kinetoplastids. Finally, we also consider the fascinating example of Perkinsela, a basal kinetoplastid and itself an endosymbiont of Paramoeba sp. (Dyková et al., Reference Dyková, Fiala, Lom and Lukeš2003; Tanifuji et al., Reference Tanifuji, Cenci, Moog, Dean, Nakayama, David, Fiala, Curtis, Sibbald, Onodera, Colp, Flegontov, Johnson-MacKinnon, McPhee, Inagaki, Hashimoto, Kelly, Gull, Lukeš and Archibald2011). Here, the evolutionary path from protist to obligate endosymbiont has been accompanied by streamlining and loss of much cell biology that defines and characterizes the Kinetoplastea (Tanifuji et al., Reference Tanifuji, Kim, Onodera, Gibeault, Dlutek, Cawthorn, Fiala, Lukeš, Greenwood and Archibald2017).

Fig. 1. Kinetoplastid phylogeny and a history of endosymbiosis. Taxa in possession of bacterial endosymbionts are highlighted in bold. Filled circles denote presence of a cytostome–cytopharynx complex in some trypanosomatid taxa; open and dashed circles denote uncertainty (as defined by an absence of data) or an unlikeliness (based on extensive, published electron microscopy studies), respectively, with regard to the presence of these structures in others; – denotes absence of a cytostome–cytopharynx from Leishmania and African trypanosome species.
Independent origins of endosymbiosis among trypanosomatids
Species belonging to four trypanosomatid genera, spanning two monophyletic groups (Fig. 1), are characterized by the presence of a bacterial endosymbiont. Phylogenetic analyses indicate Novymonas esmeraldas, the most recently characterized endosymbiont-bearing trypanosomatid isolated in Ecuador from a scentless plant bug (Niesthrea vincentii) (Kostygov et al., Reference Kostygov, Butenko, Nenarokova, Tashyreva, Flegontov, Lukeš and Yurchenko2016), is closely related to the genus Leishmania that encompasses more than 30 species of dixenous parasites found variously in tropical and sub-tropical countries across the New and Old World and include the aetiological agents of cutaneous, mucocutaneous and visceral human disease (Akhoundi et al., Reference Akhoundi, Downing, Votýpka, Kuhls, Lukeš, Cannet, Ravel, Marty, Delaunay, Kasbari, Granouillac, Gradoni and Sereno2017). Novymonas esmeraldas is considered to be monoxenous and non-pathogenic, despite its close relatedness with Leishmania. It contains a β-proteobacterial endosymbiont, Candidatus Pandoraea novymonadis (order Burkholderiales; family Burkholderiaceae) (Kostygov et al., Reference Kostygov, Dobáková, Grybchuk-Ieremenko, Váhala, Maslov, Votýpka, Lukeš and Yurchenko2017). Environmental DNA reads corresponding to 18S rRNA and trypanosomatid spliced leader RNA gene sequences point to the presence of trypanosomatid taxa very closely related to N. esmeraldas in Central Africa (Kostygov et al., Reference Kostygov, Butenko, Nenarokova, Tashyreva, Flegontov, Lukeš and Yurchenko2016), raising the question of whether such taxa also contain similar Pandoraea-related endosymbionts.
In contrast, strigomonads form a discrete monophyletic clade most closely related to the genera Wallacemonas and Sergeia and some distance removed from the leishmanias (Teixeira et al., Reference Teixeira, Borghesan, Ferreira, Santos, Takata, Campaner, Nunes, Milder, de Souza and Camargo2011; Votýpka et al., Reference Votýpka, Kostygov, Kraeva, Grybchuk-Ieremenko, Tesařová, Grybchuk, Lukeš and Yurchenko2014). In the common ancestor of the three genera-forming Strigomonadinae – Angomonas, Strigomonas and Kentomonas – an endosymbiotic association with a different member of the Burkholderiales (family Alcaligenaceae) occurred. Considered to be a more ancient relationship than the endosymbiosis occurring in N. esmeraldas, the cell cycle of the endosymbiont in strigomonads is firmly entrained within that of its host cell. The peptidoglycan and outermost layers of the endosymbiont cell envelope are absent or heavily reduced (Motta et al., Reference Motta, Catta-Preta, Schenkman, de Azevedo Martins, Miranda, de Souza and Elias1997; de Souza and Motta, Reference de Souza and Motta1999), potentially facilitating easy metabolite transfer between host and endosymbiont (discussed further in the ‘Interface with host cell biology’). Bacterial endosymbionts in Strigomonadinae are known as Candidatus Kinetoplastibacterium spp. (Alves et al., Reference Alves, Serrano, Maia da Silva, Voegtly, Matveyev, Teixeira, Camargo and Buck2013). The known distribution of strigomonads is also more cosmopolitan than that of Novymonas, as they are variously found in heteropteran and dipteran insects. Moreover, different Angomonas species have been isolated from Europe, the Americas, Africa and Australia, while Kentomonas has been isolated from Ecuador and the Philippines, and Strigomonas was encountered in different regions of the Americas (Maslov et al., Reference Maslov, Votýpka, Yurchenko and Lukeš2013; Votýpka et al., Reference Votýpka, Kostygov, Kraeva, Grybchuk-Ieremenko, Tesařová, Grybchuk, Lukeš and Yurchenko2014).
Classic hallmarks of the transition to an obligate endosymbiotic life cycle are evident in all trypanosomatid endosymbionts: a reduced GC-content (in comparison with free-living relations), reduced genome size, a paucity of mobile elements, and a reduced gene content (Table 1). Among different Candidatus Kinetoplastibacterium spp. from the strigomonads there are only slight variations in overall gene content and near-complete preservation of synteny (Alves et al., Reference Alves, Serrano, Maia da Silva, Voegtly, Matveyev, Teixeira, Camargo and Buck2013; Silva et al., Reference Silva, Kostygov, Spodareva, Butenko, Tossou, Lukeš, Yurchenko and Alves2018) indicating reductive evolution of these endosymbionts had progressed nearly to completion prior to the divergence of the last common Strigomonas/Angomonas/Kentomonas ancestor(s) or that reductive evolution of endosymbionts followed parallel trajectories in each strigomonad lineage (Alves et al., Reference Alves, Serrano, Maia da Silva, Voegtly, Matveyev, Teixeira, Camargo and Buck2013). We pick up briefly in the discussion of the ‘Interface with host cell biology’ how the reductive evolution of trypanosomatid endosymbiont gene content commonly incorporates loss of processes associated with a free-living lifestyle and perception of environmental change.
Table 1. Genome properties of trypanosomatid endosymbionts and related taxa

Ca. Pan. nov, Candidatus Pandoraea novymonadis; fl Pan. spp., free-living Pandoraea species; Ca. Kin, Candidatus Kinetoplastibacterium; Tay. equ, Taylorella equigenitalis (pathogenic bacterium closely related to Ca. Kinetoplastibacterium; Alves et al., Reference Alves, Serrano, Maia da Silva, Voegtly, Matveyev, Teixeira, Camargo and Buck2013); Ach. xyl, Achromobacter xylosoxidans (free-living bacterium closely related to Ca. Kinetoplastibacterium; Alves et al., Reference Alves, Serrano, Maia da Silva, Voegtly, Matveyev, Teixeira, Camargo and Buck2013).
Common metabolic gains in endosymbiont-containing trypanosomatids
A consequence of any endosymbiosis is conferment of new metabolic capability for the host cell. Taken to extremes, an endosymbiont's cell cycle can become entrained within that of its host and the advent of translocon-mediated protein targeting from host to endosymbiont classically marks the transition from endosymbiont to ‘organelle’ (Cavalier-Smith and Lee, Reference Cavalier-Smith and Lee1985; Theissen and Martin, Reference Theissen and Martin2006; Keeling et al., Reference Keeling, McCutcheon and Doolittle2015; McCutcheon, Reference McCutcheon2016). Among eukaryotes, the most easily recognizable products of endosymbiotic relationships are mitochondria, which conferred cytochrome-dependent oxidative phosphorylation upon an archaeal host cell of ill-defined metabolic capability (Sousa et al., Reference Sousa, Neukirchen, Allen, Lane and Martin2016; Eme et al., Reference Eme, Spang, Lombard, Stairs and Ettema2017; Zachar and Szathmáry, Reference Zachar and Szathmáry2017), and chloroplasts responsible for photosynthesis; they have evolved independently twice as the consequence of a primary endosymbiotic event (Nowack and Grossman, Reference Nowack and Grossman2012; Singer et al., Reference Singer, Poschmann, Mühlich, Valadez-Cano, Hänsch, Hüren, Rensing, Stühler and Nowack2017). These organelles were pivotal in the radiation of eukaryotic diversity with chloroplasts, notably of red algal origin, also becoming widely established in many protist lineages as consequences of secondary and tertiary endosymbiosis (Keeling, Reference Keeling2013). On a global scale, chloroplast functions remain integral to carbon cycle dynamics (Pan et al., Reference Pan, Birdsey, Fang, Houghton, Kauppi, Kurz, Phillips, Shvidenko, Lewis, Canadell, Ciais, Jackson, Pacala, McGuire, Piao, Rautiainen, Sitch and Hayes2011; Phillips and Lewis, Reference Phillips and Lewis2014; Worden et al., Reference Worden, Follows, Giovannoni, Wilken, Zimmerman and Keeling2015). At a species level, a few taxa are also secondarily photosynthetic owing to transient retention of chloroplasts (and transcriptionally active nuclei) from their algal prey (Dorrell and Howe, Reference Dorrell and Howe2012). This phenomenon is termed ‘kleptoplastidy’; such opportunistic oxygenic photosynthesis potentially confers several advantages, including aerobic respiration within anoxic environments (Esteban et al., Reference Esteban, Finlay and Clarke2009). A wide variety of other endosymbioses also exist in eukaryotic evolution that confer alternative physiological advantage(s) for the host cell as consequences of different metabolic gains, e.g. N2 fixation and N2 recycling (from waste host urea, ammonium products) occurring in termite gut-dwelling parabasalid and oxymonad flagellates and in some diatoms or, among anaerobic (non-photosynthetic) ciliates, CO2 fixation by methanogenic bacterial endosymbionts that utilize H2 produced as a metabolic end-product by the host cell (Nowack and Melkonian, Reference Nowack and Melkonian2010; Allen et al., Reference Allen, Dupont, Oborník, Horák, Nunes-Nesi, McCrow, Zheng, Johnson, Hu, Fernie and Bowler2011; Carpenter et al., Reference Carpenter, Weber, Davisson, Pett-Ridge, Haverty and Keeling2013; Tai et al., Reference Tai, Carpenter, Weber, Nalepa, Perlman and Keeling2016).
Among the Trypanosomatidae, endosymbiosis likely confers physiological advantage within nutritionally challenging environments offered by the digestive tracts of their invertebrate vectors (or hosts). However, it is neither N2 nor CO2 fixation or an ability to utilize or provide alternative carbon sources or electron acceptors for energy generation that differentiate endosymbiont-bearing trypanosomatids from other trypanosomatids. Instead, their endosymbionts render strigomonads and Novymonas autotrophic for vitamins (or cofactor precursors), amino acids, purines, and heme which are all essential nutrients in other trypanosomatids (Table 2). The curious exception is the endosymbiont from Kentomonas sorsogonicus, which is missing the haem biosynthetic pathway and the host cell is thus reliant upon an exogenous source of haem within its culture medium (Silva et al., Reference Silva, Kostygov, Spodareva, Butenko, Tossou, Lukeš, Yurchenko and Alves2018). As highlighted in Table 2, in many instances complete biosynthetic pathways are encoded within endosymbiont genomes; in other instances, metabolite exchange between endosymbiont and host is required to complete amino acid, haem or vitamin provision. For Angomonas deanei, Strigomonas culicis and S. oncopelti, predictions for autotrophy arising from genome annotations are consistent with early descriptions of minimal culture media (Newton, Reference Newton1957; Mundim et al., Reference Mundim, Roitman, Hermans and Kitajima1974; De Menezes et al., Reference De Menezes and Roitman1991).
Table 2. Metabolic gains for endosymbiont-containing trypanosomatids

RT, regular trypanosomatids; Ne, Novymonas esmeraldas; A/K/S, Angomonas/Kentomonas/Strigomonas.
a With the exception of the endosymbiont from the sole characterized Kentomonas species (K. sorsogonicus) where the haem biosynthetic pathway is absent from both host and its endosymbiont (Silva et al., Reference Silva, Kostygov, Spodareva, Butenko, Tossou, Lukeš, Yurchenko and Alves2018).
b Requires use of host cell branched-chain amino acid aminotransferase.
c Pantothenic acid synthesis utilizes enzymes from host and endosymbiont.
Whether the enhanced autotrophies of endosymbiont-containing trypanosomatids serve to widen the range of vectors that can be colonized and/or offers these trypanosomatids a competitive edge over other microbiota that may compete for the gut niche is not known. At first glance, the relative rarity of endosymbiont-containing trypanosomatids in ecological surveys argues against either of these possibilities. However, it is moot whether susceptibility to antibiotics typically applied during isolation into the culture of trypanosomatids from ecological surveys limits the frequency with which endosymbiont-bearing taxa are found. Insect digestive tracts colonized by trypanosomatids are ill-understood environments, but although they clearly provide sufficient haem, purines, vitamins of the group B and other precursors to support parasite replication in different regions of the alimentary tract, they are also unequivocally nutritionally challenging environments. Several pieces of evidence support this assertion of a nutritional ‘knife-edge’: (i) with rare exception, trypanosomatid species present (in comparison with other parasites) complex and robust metabolic networks for central energy metabolism and anabolism (notably in the extent of sterol and other lipid biosynthetic pathways) (Ginger, Reference Ginger2006; Kraeva et al., Reference Kraeva, Butenko, Hlaváčová, Kostygov, Myškova, Grybchuk, Leštinová, Votýpka, Volf, Opperdoes, Flegontov, Lukeš and Yurchenko2015; Opperdoes et al., Reference Opperdoes, Butenko, Flegontov, Yurchenko and Lukeš2016); (ii) retention in some trypanosomatids of enzymes to (a) complete biosynthetic pathways for which gut microbiota can provide initial precursors – e.g. the importance of homoserine kinase coupled to the expression of threonine synthase in tsetse-dwelling forms of the African trypanosome Trypanosoma brucei (Ong et al., Reference Ong, Lee, Patterson, Wyllie and Fairlamb2015) or (b) catabolize carbon sources likely specific to the insect vectors of some trypanosomatids – e.g. histidine in the reduviid vector of the American trypanosome T. cruzi (Berriman et al., Reference Berriman, Ghedin, Hertz-Fowler, Blandin, Renauld, Bartholomeu, Lennard, Caler, Hamlin, Haas, Böhme, Hannick, Aslett, Shallom, Marcello, Hou, Wickstead, Alsmark, Arrowsmith, Atkin, Barron, Bringaud, Brooks, Carrington, Cherevach, Chillingworth, Churcher, Clark, Corton, Cronin, Davies, Doggett, Djikeng, Feldblyum, Field, Fraser, Goodhead, Hance, Harper, Harris, Hauser, Hostetler, Ivens, Jagels, Johnson, Johnson, Jones, Kerhornou, Koo, Larke, Landfear, Larkin, Leech, Line, Lord, Macleod, Mooney, Moule, Martin, Morgan, Mungall, Norbertczak, Ormond, Pai, Peacock, Peterson, Quail, Rabbinowitsch, Rajandream, Reitter, Salzberg, Sanders, Schobel, Sharp, Simmonds, Simpson, Tallon, Turner, Tait, Tivey, Van Aken, Walker, Wanless, Wang, White, White, Whitehead, Woodward, Wortman, Adams, Embley, Gull, Ullu, Barry, Fairlamb, Opperdoes, Barrell, Donelson, Hall, Fraser, Melville and El-Sayed2005); (iii) the extensive reductive evolution of central metabolism that does occur in trypanosomatids when they become adapted to live in particularly nutrient rich environments – e.g. Phytomonas in sugar-rich plant sap (Kořený et al., Reference Kořený, Sobotka, Kovářová, Gnipová, Flegontov, Horváth, Oborník, Ayala and Lukeš2012; Porcel et al., Reference Porcel, Denoeud, Opperdoes, Noel, Madoui, Hammarton, Field, Da Silva, Couloux, Poulain, Katinka, Jabbari, Aury, Campbell, Cintron, Dickens, Docampo, Sturm, Koumandou, Fabre, Flegontov, Lukeš, Michaeli, Mottram, Szöőr, Zilberstein, Bringaud, Wincker and Dollet2014) or kinetoplast loss in mechanically – rather than tsetse-transmitted African trypanosomes (Lai et al., Reference Lai, Hashimi, Lun, Ayala and Lukeš2008). Intriguingly, the loss of respiratory complexes III and IV in Phytomonas (Nawathean and Maslov, Reference Nawathean and Maslov2000) may have helped facilitate an ability of P. françai to colonize its cyanide-rich cassava host. Comparative analysis of proteome and annotated genomes of endosymbiont-containing A. deanei and S. culcis have indicated no obvious moderation of the central metabolic networks seen in better studied Leishmania or Trypanosoma parasites (Motta et al., Reference Motta, Monteiro-Leal, de Souza, Almeida and Ferreira2013).
Interface with host cell biology I: strigomonads the slavers; Novymonas the farmer
Despite some variations in cell shape, all endosymbiont-containing trypanosomatids adopt liberform morphologies where the flagellum is not attached for an extended region to the cell body following exit from the flagellar pocket (Fig. 2).
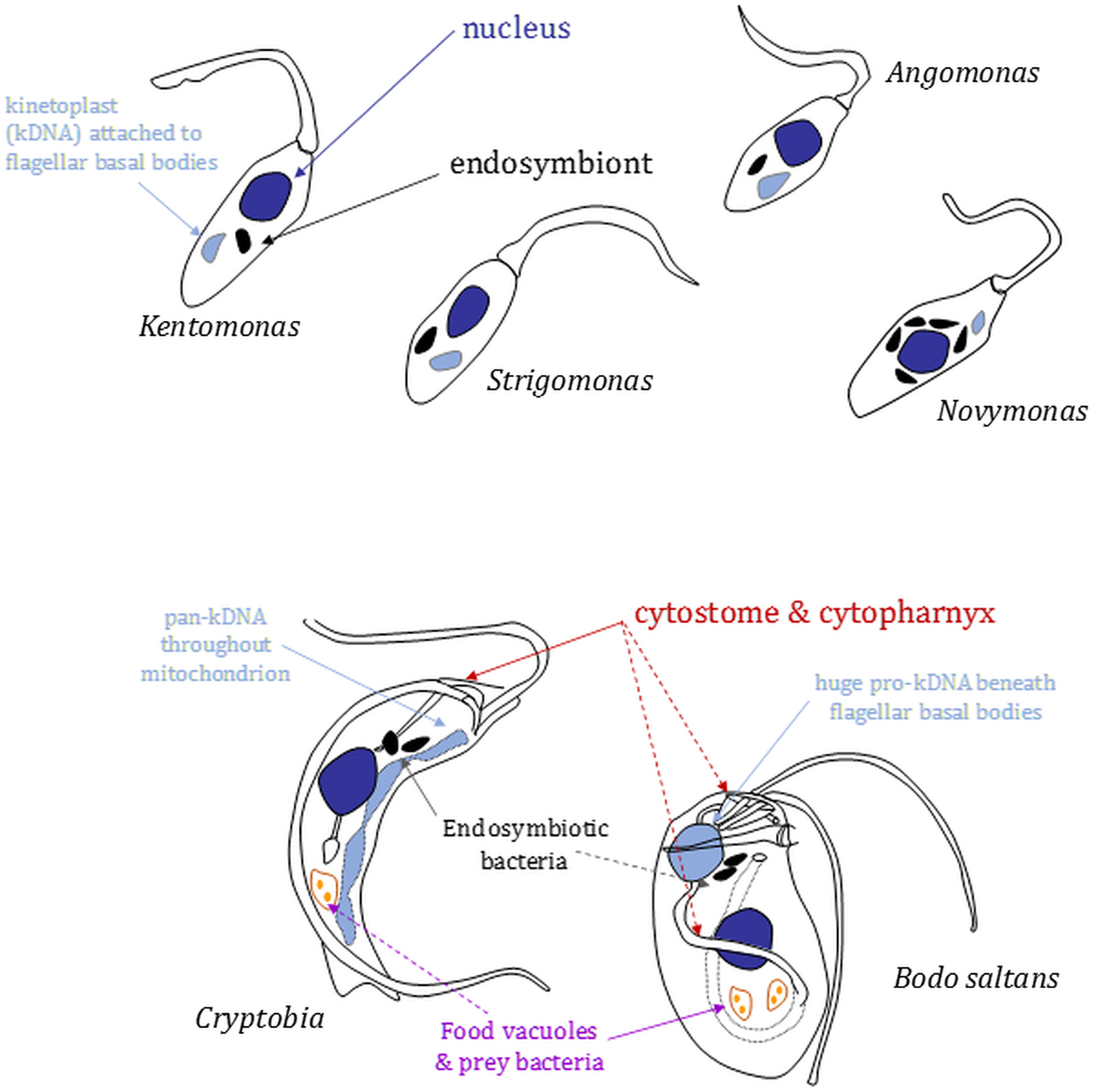
Fig. 2. Morphology and nucleus–mitochondrial genome–endosymbiont organization in endosymbiont-containing kinetoplastids. Cartoons (not to scale) are based on images shown in Kostygov et al. (Reference Kostygov, Butenko, Nenarokova, Tashyreva, Flegontov, Lukeš and Yurchenko2016), Teixeira et al. (Reference Teixeira, Borghesan, Ferreira, Santos, Takata, Campaner, Nunes, Milder, de Souza and Camargo2011) and Votýpka et al. (Reference Votýpka, Kostygov, Kraeva, Grybchuk-Ieremenko, Tesařová, Grybchuk, Lukeš and Yurchenko2014) or original drawings in Brooker (Reference Brooker1971a) and Vickerman (Reference Vickerman1977). Relative positions of several organelles discussed in the main text are shown. Shading: black, bacterial endosymbionts; dark grey, nuclei; light grey, mitochondrial genomes [kinetoplasts (kDNA) or (in Cryptobia) pan-kDNA and (in Bodo saltans) pro-kDNA].
In strigomonads, their endosymbiont is positioned proximate to the nucleus and its replication and division in the cell cycle entrained (Motta et al., Reference Motta, Martins, de Souza, Catta-Preta, Silva, Klein, de Almeida, de Lima Cunha, Ciapina, Brocchi, Colabardini, de Araujo Lima, Machado, de Almeida Soares, Probst, de Menezes, Thompson, Bartholomeu, Gradia, Pavoni, Grisard, Fantinatti-Garboggini, Marchini, Rodrigues-Luiz, Wagner, Goldman, Fietto, Elias, Goldman, Sagot, Pereira, Stoco, de Mendonça-Neto, Teixeira, Maciel, de Oliveira Mendes, Ürményi, de Souza, Schenkman and de Vasconcelos2010): endosymbiont duplication occurs early in the cell cycle preceding the host cell's discrete kinetoplast S-phase and segregation, which is coupled to flagellar basal body segregation (Ogbadoyi et al., Reference Ogbadoyi, Robinson and Gull2003); endosymbiont division is followed by movement of the endosymbionts such that each is positioned on opposite outer-faces of the nucleus; mitosis (with each nucleus associated with a single endosymbiont) and new flagellum elongation beyond the flagellar pocket exit point conclude the latter stages of the cell cycle prior to cytokinesis. Annotation of Ca. Kinetoplastibacterium genomes reveal they lack much of the machinery associated with bacterial cell division, indicating involvement from the host cell in that regard (Alves et al., Reference Alves, Serrano, Maia da Silva, Voegtly, Matveyev, Teixeira, Camargo and Buck2013; Motta et al., Reference Motta, Monteiro-Leal, de Souza, Almeida and Ferreira2013). The co-ordination of endosymbiont division within that of the host cell is illustrated further by the effect of the addition of aphidicolin, an inhibitor of eukaryotic replication DNA polymerases, or the eukaryotic translation inhibitor cycloheximide to A. deanei or S. culicis (Catta-Preta et al., Reference Catta-Preta, Brum, da Silva, Zuma, Elias, de Souza, Schenkman and Motta2015). Application of either eukaryotic growth inhibitor resulted in cessation of host cell growth and division and also blocked endosymbiont division but not endosymbiont replication. Application of aphidicolin in S. culicis additionally caused filamentation of bacteria indicating re-entry of the endosymbiont into subsequent cell cycles and continued DNA replication but without any completion of cytokinesis (Catta-Preta et al., Reference Catta-Preta, Brum, da Silva, Zuma, Elias, de Souza, Schenkman and Motta2015).
Candidatus Pandoraea novymonadis replicates more readily within the cytoplasm of its host cell (Fig. 3A and B). In multiplicative N. esmeraldas promastigotes, ~70% of the population contain between two and six endosymbionts, with 10 or more present in ~5% of cells (Kostygov et al., Reference Kostygov, Butenko, Nenarokova, Tashyreva, Flegontov, Lukeš and Yurchenko2016). Approximately 6% of Novymonas cells are aposymbiotic although the extreme difficulty in cloning such cells, the retention of intracellular bacteria in cultures since their isolation, and the significant deceleration of an aposymbiotic cell line growth as compared with wild-type highlight the importance of Ca. Pandoraea to host cell fitness (Kostygov et al., Reference Kostygov, Dobáková, Grybchuk-Ieremenko, Váhala, Maslov, Votýpka, Lukeš and Yurchenko2017). This contrasts with strigomonads where aposymbiotic populations, albeit replicating more slowly than parental lines and with increased nutritional requirements, can be readily obtained by treatment of cultures with chloramphenicol (de Souza and Motta, Reference de Souza and Motta1999). Intriguingly, studies of aposymbiotic strigomonads reveal another possible dimension to the host-endosymbiont interface with differences evident in cell surface carbohydrate composition between symbiont-containing and symbiont-lacking S. culicis cultivated in equivalent media and the indication that altered surface composition negatively influences interaction of the trypanosomatid with permissive insect hosts (Dwyer and Chang Reference Dwyer and Chang1976; Catta-Preta et al., Reference Catta-Preta, Nascimento, Garcia, Saraiva, Motta and Meyer-Fernandes2013; d'Avila-Levy et al., Reference d'Avila-Levy, Silva, Hayashi, Vermelho, Alviano, Saraiva, Branquinha and Santos2015). Significantly, culture conditions have been shown to influence the composition of the cell surface of other trypanosomatids, demonstrating common links between nutritional status and cell surface properties (Vassella et al., Reference Vassella, Den Abbeele, Bütikofer, Renggli, Furger, Brun and Roditi2000; Morris et al., Reference Morris, Wang, Drew and Englund2002).

Fig. 3. Electron microscopy of the endosymbiont–host cell association and cell form in Novymonas and Kentomonas. (A and B) Longitudinal sections through N. esmeraldas promastigotes showing the presence of multiple endosymbiont profiles (e). Also highlighted are the kinetoplast (K), nucleus (N) and cross-sections through the mitochondrion (m). (C) Longitudinal section through a Kentomonas sorsogonicus choanomastigote illustrating (i) a dividing bacterial endosymbiont and (ii) mitochondrial hypertrophy and loss of typical microtubule spacing within the sub-pellicular array. (D) Sessile N. esmeraldas choanomastigote attached to the substrate surface via a modified flagellum (asterisk). Inset, the modified flagellum of a sessile choanomastigote revealing a possible open collar structure to the flagellar pocket exit point. Scale bars (A) and (B) 2 µm; (C) 1 µm; (D) 2 µm (inset, 400 nm). Images in (D) are reproduced from Kostygov et al. (Reference Kostygov, Butenko, Nenarokova, Tashyreva, Flegontov, Lukeš and Yurchenko2016) under the terms of a Creative Commons Attribution-Noncommercial-ShareAlike 3.0 Unported licence.
Fusion of Novymonas lysosomes with Ca. P. novymonadis provides an indication that the host ‘farms’ its endosymbiont, presumably taking amino acids, haem, purines and other molecules liberated in lysosomes to satisfy dietary requirements. In agreement with the ‘lax’ control on endosymbiont multiplication evident in Novymonas, Ca. P. novymonadis retains more genes associated with bacterial cell division than Ca. Kinetoplastibacterium (Kostygov et al., Reference Kostygov, Dobáková, Grybchuk-Ieremenko, Váhala, Maslov, Votýpka, Lukeš and Yurchenko2017). However, other findings from Ca. P. novymonadis genome annotation point to a well-established host–endosymbiont relationship and provide a note of caution for any assumption of how readily Ca. P. novymonadis might multiply free from the host cell in different, commonly used bacterial growth media. For instance, cellular characteristics associated with perception and response to environmental change are either absent (genes for pilus and flagellum assemblies, ‘wsp’ chemotaxis proteins, ‘pel’ proteins involved in biofilm formation) or minimalized (two-component signalling). There is also a drastic reduction in the number of nutrient transporters/exporters present, including members of ABC-transporter and major facilitator superfamilies and in the ability of Ca. P. novymonadis to catabolize diverse carbon sources in comparison with free-living Pandoraea (Fig. 4).

Fig. 4. In silico annotated proteomes illustrate the reductive evolution of Ca. Pandoraea novymonadis and Ca. Kinetoplastibacterium. Predicted protein repertoires for Ca. P, novymonadis, 5 Ca. Kinetoplastibacterium spp. and 11 free-living Pandoraea species (Kostygov et al., Reference Kostygov, Dobáková, Grybchuk-Ieremenko, Váhala, Maslov, Votýpka, Lukeš and Yurchenko2017) were analysed according to within the KEGG Orthology (KO). 2728 KO functions were analysed. For Ca. Kinetoplastibacterium spp. and Pandoraea spp. annotation of gene products in 3 or 5 genomes, respectively, were required for inclusion in the chart shown. Known nearest free-living relatives of Ca. Kinetoplastibacterium are evolutionarily more distant than for Ca. P. novymonadis, and were not therefore included in the analysis although we note the closest Ca. Kinetoplastibacterium free-living relative, A. xylosoxidans, is more gene-rich than free-living Pandoraea spp. (Table 2). Individual gene products were scored once and appear in only one of the following categories. Central metabolism: category 1, carbohydrate usage (including lipopolysaccharide and peptidoglycan assembly); 2, amino acid catabolism; 3, amino acid biosynthesis (including glycolysis); 4, fatty acid and terpenoid metabolism; 5, inositol phosphate and glycerophospholipid metabolism; 6 butanoate and propanoate metabolism; 7, pyruvate, glyoxylate, and dicarboxylate metabolism; 8, degradation of aromatics; 9, pentose phosphate and antioxidant metabolism; 10, Krebs cycle; 11, respiration and oxidative phosphorylation. Accessory metabolism: 12, porphyrin metabolism; 13, miscellaneous (including carbon fixation, sulphur and methane metabolism, urease); 14, vitamin and cofactor biosynthesis; 15, transporters and ATPases. Information processing: 16, replication and DNA repair; 17, purine and pyrimidine metabolism (including tRNA processing and core transcription); 18, ribosome and translation; 19, chaperones. Environmental responses: 20, two-component signaling, transcriptional regulation, quorum sensing and phosphate metabolism; 21, cell division; 22, secondary metabolism and antibiotic defence/attack; 23, flagellum, pilus, biofilm formation.
However, metabolic dependencies in endosymbiotic relationships go both ways. In the trypanosomatid examples, owing in large part to the close proximity of strigomonad endosymbionts to host cell mitochondria and glycosomes, strigomonads have for many years been considered to provide ATP to their intracellular partners (see Loyola-Machado et al., Reference Loyola-Machado, Azevedo-Martins, Catta-Preta, de Souza, Galina and Motta2017 for recent consideration of this topic). This assertion is supported by the paucity of options for efficient oxidative phosphorylation by Ca. Kinetoplastibacterium spp.
Genomes of both Ca. Kinetoplastibacterium and Ca. P. novymonadis contain genes for nuo-type NADH:ubiquinone oxidoreductases (Kostygov et al., Reference Kostygov, Dobáková, Grybchuk-Ieremenko, Váhala, Maslov, Votýpka, Lukeš and Yurchenko2017), but in the former its electron transport chain is truncated to a cytochrome bd terminal oxidase for transfer of electrons from ubiquinone to O2 – the type of terminal oxidase favoured by numerous bacteria, including Escherichia coli under low O2 availability. In contrast to Ca. Kinetoplastibacterium spp., however, whilst the carbon source(s) utilized by Novymonas endosymbionts remains enigmatic – fructose, common in the diet of plant-feeding insects, is the most likely carbon source (Kostygov et al., Reference Kostygov, Dobáková, Grybchuk-Ieremenko, Váhala, Maslov, Votýpka, Lukeš and Yurchenko2017) – the novymonad endosymbiont appears more self-sufficient for energy generation. Perhaps as a consequence of their greater autonomy with regard to their rate of cell division, and thus a greater need for intra-symbiont ATP generation, Ca. P. novymonadis retains a more expansive electron transport chain. Here the metabolism includes a capacity for oxidative phosphorylation from c-type cytochrome-dependent respiration.
Currently, the least explored facet of the interface from host to endosymbiont is the degree to which the host cell targets nuclear-encoded proteins to the symbiont. One example is known for A. deanei (Morales et al., Reference Morales, Kokkori, Weidauer, Chapman, Goltsman, Rokhsar, Grossman and Nowack2016b), but this is a long way short of the number of host-targeted proteins that might be required to question whether trypanosomatid endosymbionts begin to blur boundaries between endosymbiont and organelles.
Interface with host cell biology II: symbiont acquisition by closed-mouth, osmotrophic trypanosomatids – how?
In contrast to phagotrophic bodonids and other free-living kinetoplastids, trypanosomatids are obligate osmotrophs. A robust sub-pellicular mono-layer of microtubules cross-linked to one another and the over-laying plasma membrane provides a corset that defines characteristic trypanosomatid cell morphologies and prevents general endocytosis or membrane invagination across the cell surface. Membrane invagination occurs only at points where the sub-pellicular corset is absent which, in well-studied African trypanosomes and Leishmania, is where the flagellar pocket forms around the single flagellum emerging from the cell body. In these trypanosomatids, the flagellar pocket is the site of endo- and exocytic traffic (Field and Carrington, Reference Field and Carrington2009). At the flagellum exit point, an essential collar marks the flagellar pocket boundary (Bonhivers et al., Reference Bonhivers, Nowacki, Landrein and Robinson2008) limiting the size and rate of macromolecular traffic into the pocket lumen (Gadelha et al., Reference Gadelha, Wickstead, de Souza, Gull and Cunha-e-Silva2009). Given these constraints, how, following radiation of various trypanosomatid lineages, have trypanosomatid–endosymbiont associations occurred on at least two occasions?
Several possibilities can explain the conundrum of how Novymonas and a strigomonad ancestor acquired their respective bacterial endosymbionts. Conserved in free-living kinetoplastids and present in some trypanosomatids (Brooker, Reference Brooker1971a, Reference Brooker1971b; Brugerolle et al., Reference Brugerolle, Lom, Nohynkova and Joyon1979; Attias et al., Reference Attias, Vommaro and de Souza1996; Alcantara et al., Reference Alcantara, Vidal, de Souza and Cunha-E-Silva2017; Skalický et al., Reference Skalický, Dobáková, Wheeler, Tesařová, Flegontov, Jirsová, Votýpka, Yurchenko, Ayala and Lukeš2017) is a cytostome–cytopharynx complex, sitting in close proximity to the flagellar pocket (Figs 1 and 5). In T. cruzi (Porto-Carreiro et al., Reference Porto-Carreiro, Attias, Miranda, De Souza and Cunha-e-Silva2000) and apparently in Crithidia fasciculata (Brooker, Reference Brooker1971b) the cytostome is a site of endo- and pinocytosis. In free-living kinetoplastids, the cytostome leading to the cytopharynx, in conjunction with the anterior flagellum, is used for phagotrophic feeding on bacterial prey. Early microscopy analyses indicate extensive distension of the feeding apparatus in order to ingest large prey (Brooker, Reference Brooker1971a; Burzell, Reference Burzell1973, Reference Burzell1975). Enzymatic machinery necessary for digestion of complex macromolecular structures from live prey is considered to have been lost at an early point following divergence of the last common trypanosomatid ancestor (Skalický et al., Reference Skalický, Dobáková, Wheeler, Tesařová, Flegontov, Jirsová, Votýpka, Yurchenko, Ayala and Lukeš2017), coincident with the advent of obligate osmotrophy but also indicating that fortuitous uptake of a bacterium by a cytostome-bearing trypanosomatid would not necessarily be followed by its digestion.

Fig. 5. Relative positions of flagella, cytostome, cytopharynx and other cellular features in free-living Bodo and Cryptobia kinetoplastids. Images were adapted from original drawings in Figs 4–6 from Brugerolle et al. (Reference Brugerolle, Lom, Nohynkova and Joyon1979). Abbreviations (translated from the original French): Cr, oral ridge; Fas, ‘microtubule fibre’ associated with the ‘striatal plaque’; Fd, ‘dorsal fibre’; Fr, recurrent flagellum; Fv, ‘ventral fibre’; Fa, anterior flagellum; G, Golgi; K, kintetoplast; M, mitochondrion; mb, microbodies; mtr, ‘reinforced microtubules; N, nucleus; Pf, flagellar pocket; Vc, contractile vacuole; Vd, food vacuole.
Although clearly absent from African trypanosomes and Leishmania (Skalický et al., Reference Skalický, Dobáková, Wheeler, Tesařová, Flegontov, Jirsová, Votýpka, Yurchenko, Ayala and Lukeš2017), a paucity of data cannot yet allow insight into how often and when the cytostome–cytopharynx was lost during trypanosomatid evolution. Whilst this organelle complex has never been seen from detailed ultrastructural analyses of extant strigomonads (Bombaça et al., Reference Bombaça, Dias, Ennes-Vidal, Garcia-Gomes, Sorgine, d'Avila-Levy and Menna-Barreto2017; Loyola-Machado et al., Reference Loyola-Machado, Azevedo-Martins, Catta-Preta, de Souza, Galina and Motta2017) or analysis of Phytomonas sp. (e.g. Postell and McGhee, Reference Postell and McGhee1981; Milder et al., Reference Milder, Camargo and Freymullar1990) and functionality of the Crithidia ‘cytostome’ has not, to our knowledge been revisited since the early 1970s, the critical questions are whether an ancestral cytostome was present and could have played a role in endosymbiont uptake by strigomonad and/or novymonad ancestors. A cytostome–cytopharynx is retained in the basal trypanosomatid Paratrypanosoma confusum (Skalický et al., Reference Skalický, Dobáková, Wheeler, Tesařová, Flegontov, Jirsová, Votýpka, Yurchenko, Ayala and Lukeš2017); coupled to the monophyly of the trypanosomes, plus the relatively close relationship between the leishmanias and C. fasciculata, the pattern of organelle degeneration and thence loss was likely complex. The observation that cytostome–cytopharynx assembly in T. cruzi is stage-regulated (Vidal et al., Reference Vidal, Alcantara, de Souza and Cunha-E-Silva2016) also leaves open the possibility of a cryptic or hidden cytostome in other extant trypanosomatids. Thus, cell entry via a cytostome is a plausible route for the acquisition of Novymonas or strigomonad endosymbionts.
To consider alternative acquisition routes, a hypertrophied mitochondrion is a diagnostic trait for the Strigomonadinae and its invasion of the spacing between sub-pellicular microtubules (Fig. 3C) is often considered to be a consequence of endosymbiosis with the ATP requirements of the endosymbiont driving mitochondrial expansion and an increased rate of energy generation by the host cell. Looser organization of the kinetoplast, relative to other trypanosomatids, is another strigomonad-specific characteristic (Teixeira et al., Reference Teixeira, Borghesan, Ferreira, Santos, Takata, Campaner, Nunes, Milder, de Souza and Camargo2011; Votýpka et al., Reference Votýpka, Kostygov, Kraeva, Grybchuk-Ieremenko, Tesařová, Grybchuk, Lukeš and Yurchenko2014), conceivably facilitates high rates of mitochondrial gene expression, and, thus, potentially an enhanced capacity for oxidative phosphorylation relative to some other trypanosomatids (careful, cross-species quantitative assessment of metabolic rate as a function of growth rate(s) under equivalent conditions will be necessary to determine if this is the case). Considered less often, however, is the possibility that mitochondrial hypertrophy and/or disruption of sub-pellicular microtubule spacing preceded endosymbiont acquisition. In this instance, a release of constraints on plasma membrane invagination would facilitate another route for endosymbiont uptake in the ancestor of the Strigomonadinae.
Looking further at the influence(s) of mitochondrial hypertrophy, rather than the endosymbiont itself might exert on host cell biology, then another strigomonad synapomorphy is the extensive reduction of paraflagellar rod (PFR) architecture. This results in a vestigial structure extended along only the proximal third of the axoneme (Gadelha et al., Reference Gadelha, Rothery, Morphew, McIntosh, Severs and Gull2006). Reductive PFR evolution was driven, at least in part, by the loss of genes encoding the major PFR2 protein. The extreme alteration of PFR form is intriguing not least because of the essentiality of this flagellar structure in other trypanosomatids (Maga et al., Reference Maga, Sherwin, Francis, Gull and LeBowitz1999; Ginger et al., Reference Ginger, Collingridge, Brown, Sproat, Shaw and Gull2013; Lander et al., Reference Lander, Li, Niyogi and Docampo2015). If the view that the PFR provides an important function in maintaining intraflagellar nucleotide homeostasis is correct (Pullen et al., Reference Pullen, Ginger, Gaskell and Gull2004; Ginger et al., Reference Ginger, Portman and McKean2008), then a significant increase to the efficiency of mitochondrial ATP production in strigomonads could have provided a selective driver for the enigmatic reduction of PFR form seen in this trypanosomatid group.
Mitochondrial hypertrophy is not so evident in Novymonas with sub-pellicular microtubules having a spacing reminiscent of that found in most trypanosomatids. Acquisition of its symbiont is thus unlikely to have occurred via invagination of the plasma membrane. Sessile N. esmeraldas choanomastigotes, however, attach to surfaces via their flagellum and attached in this way exhibit a drastically altered flagellum structure (Fig. 3D) reminiscent of the flagellum surface attachment remodelling seen also in P. confusum (Skalický et al., Reference Skalický, Dobáková, Wheeler, Tesařová, Flegontov, Jirsová, Votýpka, Yurchenko, Ayala and Lukeš2017). In scanning electron micrographs of detached N. esmeraldas choanomastigotes (Fig. 3D; inset) the altered flagellum morphology hints at a more open flagellar pocket collar through which a flagellum membrane-attached bacterium could putatively be ingested.
Endosymbioses within free-living phagotrophic kinetoplastids
There is currently sparse data with regard to endosymbionts and their role(s) in free-living phagotrophic kinetoplastids. This is not surprising given that attention to their molecular cell biology using modern approaches is only recently forthcoming (Gomaa et al., Reference Gomaa, Garcia, Delaney, Girguis, Buie and Edgcomb2017). However, constraints that leave the conundrum of how at least two trypanosomatids acquired their endosymbionts – arrayed sub-pellicular microtubules; a closed flagellar pocket – are not conspicuous among free-living kinetoplastids. Plus, there are likely significant insights to be made with regard to niche adaptation and exploitation; anoxic environments provide an obvious example with kinetoplastids being one of the few protist groups for which there is only limited evidence of adaptation (Priya et al., Reference Priya, Haridas and Manilal2008). The current lack of known anaerobic kinetoplastids contrasts with observations of obligately aerobic metabolism in trypanosomatid and Bodo saltans genomes that nonetheless showcases several anaerobic hallmarks (Michels et al., Reference Michels, Chevalier, Opperdoes, Rider and Rigden1997; Annoura et al., Reference Annoura, Nara, Makiuchi, Hashimoto and Aoki2005; Opperdoes et al., Reference Opperdoes, Butenko, Flegontov, Yurchenko and Lukeš2016).
Surveying the literature indicates that the presence of endosymbiotic bacteria is not an obligate characteristic of free-living kinetoplastids, e.g. an absence from Rhynchomonas metabolita (Burzell, Reference Burzell1973). When present, however, endosymbionts are found in the anterior region of the cytoplasm or in close proximity to the nucleus of other kinetoplastids, albeit far from the posterior cell region that tends to be dominated by food vacuoles containing bacteria ingested via the cytostome–cytopharynx (Fig. 2) (Brooker Reference Brooker1971a; Burzell, Reference Burzell1975; Vickerman, Reference Vickerman1977). Likely bacterial epibionts have been noted on the surface of Cryptobia vaginalis (Vickerman, Reference Vickerman1977) and in others endosymbiont multiplication keeps pace with host cell division and a reduced peptidoglycan layer of the endosymbiont cell wall is in evidence, again indicative of the establishment of long-term endosymbioses.
Only distantly related to the kinetoplatids, but nonetheless of interest, another clade of euglenozoans – Symbiontida – is characterized by a dense layer of ectosymbiotic bacteria, present on their surface. This poorly studied group of flagellates, consisting of only three known species, inhabit low-oxygen sea environments. The function of the ectosymbionts is not known (Yubuki et al., Reference Yubuki, Simpson and Leander2013).
Perkinsela: the enslaved kinetoplastid
The ancestor of Perkinsela, which is most closely related to the fish ectoparasite Ichthyobodo, is thought to have diverged early in kinetoplastid evolution (Fig. 1). Extant Perkinsela is an obligate endosymbiont of lobose amoebae genus Paramoeba (phylum Amoebozoa), which are pathogenic to a variety of marine animals, including farmed fish. GC-content in Perkinsela is not reduced in comparison with other sampled kinetoplastids (Tanifuji et al., Reference Tanifuji, Kim, Onodera, Gibeault, Dlutek, Cawthorn, Fiala, Lukeš, Greenwood and Archibald2017) even though the Perkinsella–Parmoeba endosymbiosis is a long time established association (Sibbald et al., Reference Sibbald, Cenci, Colp, Eglit, O'Lelly and Archibald2017). In contrast, gene content of Perkinsela is significantly reduced in comparison with free-living B. saltans and parasitic trypanosomatids – 5252 protein-coding genes in Perkinsela vs 18 943 genes in B. saltans; 6381 in Phytomonas sp.; 9068 in T. brucei (although this includes expansion of its critical antigenic variant surface glycoprotein gene repertoire); and 8272 genes in Leishmania major. This reductive evolution reflects the secondary loss of much of the cell biology that characterizes kinetoplastid cell form (Tanifuji et al., Reference Tanifuji, Kim, Onodera, Gibeault, Dlutek, Cawthorn, Fiala, Lukeš, Greenwood and Archibald2017). Ichthyobodo, in contrast, displays the biflagellate morphology typical of non-trypanosomatid kinetoplastids (Grassé, Reference Grassé and Grassé1952).
Lost from the genome of Perkinsela are all the genes required for basal body/flagellum assembly and architecture, together with an absence of genes encoding homologues of trypanosomatid cytoskeletal proteins. The absence of sub-pellicular microtubules relieves the constraints on the surface siting of endocytosis and leaves the endosymbiont able to readily ingest cytoplasm from the host (Tanifuji et al., Reference Tanifuji, Kim, Onodera, Gibeault, Dlutek, Cawthorn, Fiala, Lukeš, Greenwood and Archibald2017). Metabolism of Perkinsela is also minimized: glycolysis occurs but obvious metabolic routes from pyruvate to acetyl-CoA are lacking; a truncated Krebs’ cycle running from α-ketoglutarate to oxaloacetate likely uses a (host-derived) glutamate carbon source and provides electrons to fuel a mitochondrial respiratory chain truncated by the loss of complex I (NADH:ubiquinone oxidoreductase). It is likely that the benign environment offered by the Paramoeba host, with respect to carbon provision, facilitates the reductive evolution of intermediary metabolism. An absence of sterol metabolism potentially reflects either absence of sterol from endosymbiont membranes (similar to a few other eukaryotes) or a possibility that the host provides an easy availability of the ergosta- and stigmasta-type sterols found in other amoebozoans and trypanosomatids (Raederstorff and Rohmer, Reference Raederstorff and Rohmer1985; Nes et al., Reference Nes, Norton, Crumley, Madigan and Katz1990; Roberts et al., Reference Roberts, McLeod, Rice, Ginger, Chance and Goad2003). A lack of sugar nucleotide biosynthesis possibly indicates a reduced requirement for protein glycosylation or limited need for investment in a protective cell surface glycocalyx.
Benefits arising from an intracellular lifestyle for Perkinsela are clear, although this is not to suggest that the lifestyle is lazy: the kinetoplastid makes a huge investment in RNA editing, perhaps as a consequence of the neutral evolutionary ratchet discussed by Lukeš (Reference Lukeš, Guilbride, Votýpka, Zíková, Benne and Englund2011), for the expression of the six (essential) respiratory chain components encoded on the mitochondrial genome (David et al., Reference David, Flegontov, Gerasimov, Tanifuji, Hashimi, Logacheva, Maruyama, Onodera, Gray, Archibald and Lukeš2015) and the nuclear genome hints at the presence of a sexual cycle that is perhaps integrated within that of its host (Tanifuji et al., Reference Tanifuji, Kim, Onodera, Gibeault, Dlutek, Cawthorn, Fiala, Lukeš, Greenwood and Archibald2017).
Apart from Paramoeba and Perkinsela, all known endosymbioses involving only eukaryotes bring the provision of photosynthesis to the host partner (David et al., Reference David, Flegontov, Gerasimov, Tanifuji, Hashimi, Logacheva, Maruyama, Onodera, Gray, Archibald and Lukeš2015). What Parameoba derives from its unusual endosymbiont is currently a mystery and a source only for speculation.
Concluding remarks
Endosymbiosis is a feature of kinetoplastid evolution. Several case examples provide tractable opportunities to understand how, at the host–endosymbiont interface, long-lasting endosymbiotic relationships become established in microbial eukaryotes and leave other questions that will likely be more challenging to address. Of the latter, until more robust culture systems for Paramoeba are forthcoming, it will be difficult to establish what Perkinsela provides for its host. Similarly, without relevant traits being revealed in continuing surveys of trypanosomatid diversity, the chronology and interplay between endosymbiont acquisition, mitochondrial hypertrophy, altered kinetoplast structure and PFR reduction cannot realistically be addressed. However, we now work in an era of easy next-generation sequencing. Thus, paralleling combined genomic, transcriptomic and proteomic studies of environmentally sourced protists such as the breviate Lenisia limosa (Hamann et al., Reference Hamann, Gruber-Vodicka, Kleiner, Tegetmeyer, Riedel, Littmann, Chen, Milucka, Viehweger, Becker, Dong, Stairs, Hinrichs, Brown, Roger and Strous2016), there is much scope to reveal what endosymbionts (and epibiotic bacteria) contribute to free-living kinetoplastid hosts. Similarly, and in contrast to many other examples of protists with endosymbionts, the tractability of trypanosomatids towards genetic manipulation (Morales et al., Reference Morales, Kokkori, Weidauer, Chapman, Goltsman, Rokhsar, Grossman and Nowack2016b) leaves huge opportunity to dissect at a molecular level the regulatory influences on endosymbiont growth and division in strigomonads and Novymonas. Genetic tractability also provides the means to probe the extent to which protein targeting from host-to-symbiont (and perhaps vice versa) also eclipses the biology of trypanosomatid–endosymbiont associations.
Financial support
This work received support from the Czech Grant Agency (16-18699S to J.L.) and the European Regional Development Fund [project CePaViP (OPVVV16_019/0000759 to J. L. and V. Y.)].
Conflict of interest
None.
Ethical standards
Not applicable.










