INTRODUCTION
Dengue fever is caused by a virus that belongs to the Flavivirus genus and is endemic in tropical and subtropical areas. A study applying cartographic methods estimated an annual occurrence of 390 million dengue viral infections globally [Reference Bhatt1]. This estimation is more than three times higher than the numbers pointed previously by the World Health Organization, which ranged from 50 to 100 million cases [Reference Bhatt1]. The study using cartographic methods also estimated 96 million symptomatic dengue fever cases per year in the world [Reference Bhatt1]. The dengue virus complex is composed by four serotypes. There are genetic variants within each serotype constituting phylogenetic clusters, denominated genotypes, which differ in geographical distribution [Reference Pinho2], virulence and transmissibility [Reference Rico-Hesse3]. The genome of this ribonucleic acid (RNA) virus has about 10 700 nucleotides, encoding three structural and seven non-structural proteins [Reference Kuhn4] with considerable genetic variation. This variation is a consequence of mutations caused by RNA polymerase [Reference Twiddy, Holmes and Rambaut5] and a result of natural recombination [Reference Chen6]. The adaptive potential of the virus favours the territorial expansion of the disease [Reference Mendez7].
Dengue fever re-emerged in Brazil in the 1980s, when dengue virus serotype 1 and dengue virus serotype 4 (DENV-4) were detected in the country [Reference Rodriguez-Barraquer8]. At that time, the circulation of DENV-4 was restricted to the northern states and remained detectable during only a few months. After more than 20 years without DENV-4 detection in the country, this serotype was observed again in 2010 in Roraima and subsequently spread to other locations in Brazil [Reference Temporão9].
Dengue fever cases have been reported in Espírito Santo state since 1995. The first detection of DENV-4 occurred only in 2012 when the other three serotypes were already established in the state. According to the Health Department of Espírito Santo state, in the first 2 years after the detection of DENV-4, 107963 suspected dengue fever cases were reported in the state: 82 928 in 2013 and 25 035 in 2014 [10]. The epidemic in 2013 accounted for the highest number of dengue fever incident cases in the state history and amounted to 1595 hospitalisations [11], 74 cases of dengue haemorrhagic fever and 12 deaths [12].
Genetic characterisation [Reference Mendez7] and phylogenetic reconstructions of viruses circulating in a given population help to elucidate the role of human movements in dengue viral spread [Reference Koo13] and contribute to understanding epidemic patterns of this disease [Reference Mendez7]. This study presents the first phylogeny of DENV-4 circulating in Espírito Santo state, with samples collected during an unprecedented epidemic related to this serotype.
METHODS
Samples
Blood samples were collected from 56 patients with suspected dengue viral infection during the viraemic phase (up to 3 days after the onset of symptoms), who consulted the public health units located in Vitória, the capital of Espírito Santo state, Brazil, in 2013 and 2014. Whole blood (2·5 ml) was collected and stored at −80 °C until the beginning of the extraction process.
Molecular analysis
The QIAamp RNA Blood Mini Kit™ produced by QIAGEN© was used for the extraction of the viral RNA from the whole blood, following the instructions of the manufacturer. The High-Capacity cDNA Reverse Transcription Kit™ (Thermo Fisher Scientific©) was used to synthesise the complementary deoxyribonucleic acid, according to the manufacturer's instruction. Qualitative reverse-transcriptase-polymerase chain reaction (PCR) assay [Reference Felix14] was used to confirm dengue viral infection in the samples. Fourteen samples were positive for the presence of dengue virus RNA, but six of them presented a low viral load, which consequently impaired the successful amplification by conventional PCR. The primers used to amplify a fragment of the envelope gene of 1300 nucleotides (position 564–1864 of the DENV-4 genome – reference ID JN559741) were ENVD4_Fw – CACGTATAAATGCCCCCTACTGGTC and ENVD4_Rv – GCTGTGTTTCTGCCATCTCTTTGTC. The PCR was prepared to a final volume of 50 µl containing 10 µl of the template, 10× PCR buffer, 1·5 mM of MgCl2, 200 mM of dNTPs, 1 mM of each primer and 1 U of platinum Taq DNA polymerase (Invitrogen©). Cycling condition for PCR was as follows: 45 cycles of 94 °C for 30 s, 55 °C for 1 min and 72 °C for 1 min. Seven minutes were taken for the final extension. Amplicons were purified through GFX purification kit (GE Healthcare©) according to the manufacturer instructions. They were sequenced directly by Sanger methodology, using the same primer pairs of the amplification reaction.
Phylogenetic and coalescent reconstruction
Reads were inspected for quality, trimmed and assembled by CLC genomic Workbench v.6 (http://www.qiagenbioinformatics. com/products/clc-genomics-workbench/). Consensus sequences were aligned together with reference sequences retrieved from GenBank (http://www.ncbi.nlm.nih.gov/genbank/) in the software Clustal X [Reference Thompson15]. An initial dataset (n = 93) including sequences from Espírito Santo state (n = 8) and reference sequences from all four serotypes (n = 85) was used to confirm the serotype. A second dataset including samples from Espírito Santo state (n = 8) and reference sequences (n = 70) of different DENV-4 genotypes was created (n = 78) and used to reconstruct the phylogenetic tree. Sequences of DENV-4 circulating in São Paulo in 2015 (n = 2) were also included in order to highlight the genetic variability of strains recently circulating in Brazil. The GTR + I + G substitution model was found to be the best model for both datasets according to jModelTest [Reference Posada16]. Phylogenetic estimates were made using the Bayesian Markov Chain Monte Carlo (MCMC) framework implemented in BEAST v1.8.3 [Reference Drummond and Rambaut17]. Convergence of parameters during the MCMC was inspected with Tracer v.1.6 [Reference Drummond and Rambaut17], with uncertainties addressed as 95% highest probability density intervals. Twenty million chains were sufficient to achieve convergence on all parameters (ESS.200). The trees were sampled at each 2000 steps, resulting in a final file of 10 000 trees, summarised in a maximum clade credibility tree using TreeAnotator (part of the BEAST package), and visualised in FigTree v.1.4.2 (http://beast.bio.ed.ac.uk/FigTree). The least constrained Bayesian Skyline (BSL) coalescent prior was used to estimate the rate of nucleotide substitution and the Time to the Most Recent Common Ancestor (TMRCA). The BSL was performed under relaxed (exponential and uncorrelated lognormal) molecular clocks. The molecular clock that best fit the data was evaluated by Bayes factor (BF) comparison.
Compliance with ethical standards
The Research Ethics Committee of the Federal University of Espírito Santo (opinion number 881909) and the Research Ethics Committee of the University of Munich (opinion number 231-15) approved the research protocol. All participants signed an informed consent form.
RESULTS
Eight partial envelope gene samples were successfully amplified and sequenced. The reconstructed phylogenetic tree shows that isolates from Espírito Santo state belong to the genotype II [Reference Lanciotti, Gubler and Trent18]. DENV-4 viruses circulating in 2013 and 2014 were closely related to viruses isolated in 2012 in Mato Grosso state and in 2015 in São Paulo state, with a posterior probability value of 0·98 for this clade. According to the MCMC estimates, DENV-4 evolves at a rate of 1·1E3, which was close to the previous estimates for this serotype [Reference Twiddy, Holmes and Rambaut5]. Using this rate of nucleotide substitution estimated under the relaxed uncorrelated lognormal molecular clock (BF = 4·96), DENV-4 genotype II was introduced to the American continent between 1973 and 1981 (Fig. 1, node A). The strains circulating in Puerto Rico during that period generated a lineage that spread to the other Caribbean and South American countries. More than 20 years later, in the 2000s, at least two distinct introductions of DENV-4 genotype II occurred in Brazil. One was restricted to Pará state between 2004 and 2010 (Fig. 1, node B), with possible Caribbean origin. The other introduction occurred between 2004 and 2008 (Fig. 1, node C) in the region of Amazonas state and Roraima state, with probable Venezuelan and Colombian origins. From Roraima state, the virus spread to other Brazilian states, reaching the locations described within the clade, such as Espírito Santo state, between 2010 and 2012 (Fig. 1, node D). Figure 2 presents a map illustrating the possible origins of the strains circulating in the Brazilian states included in the study.
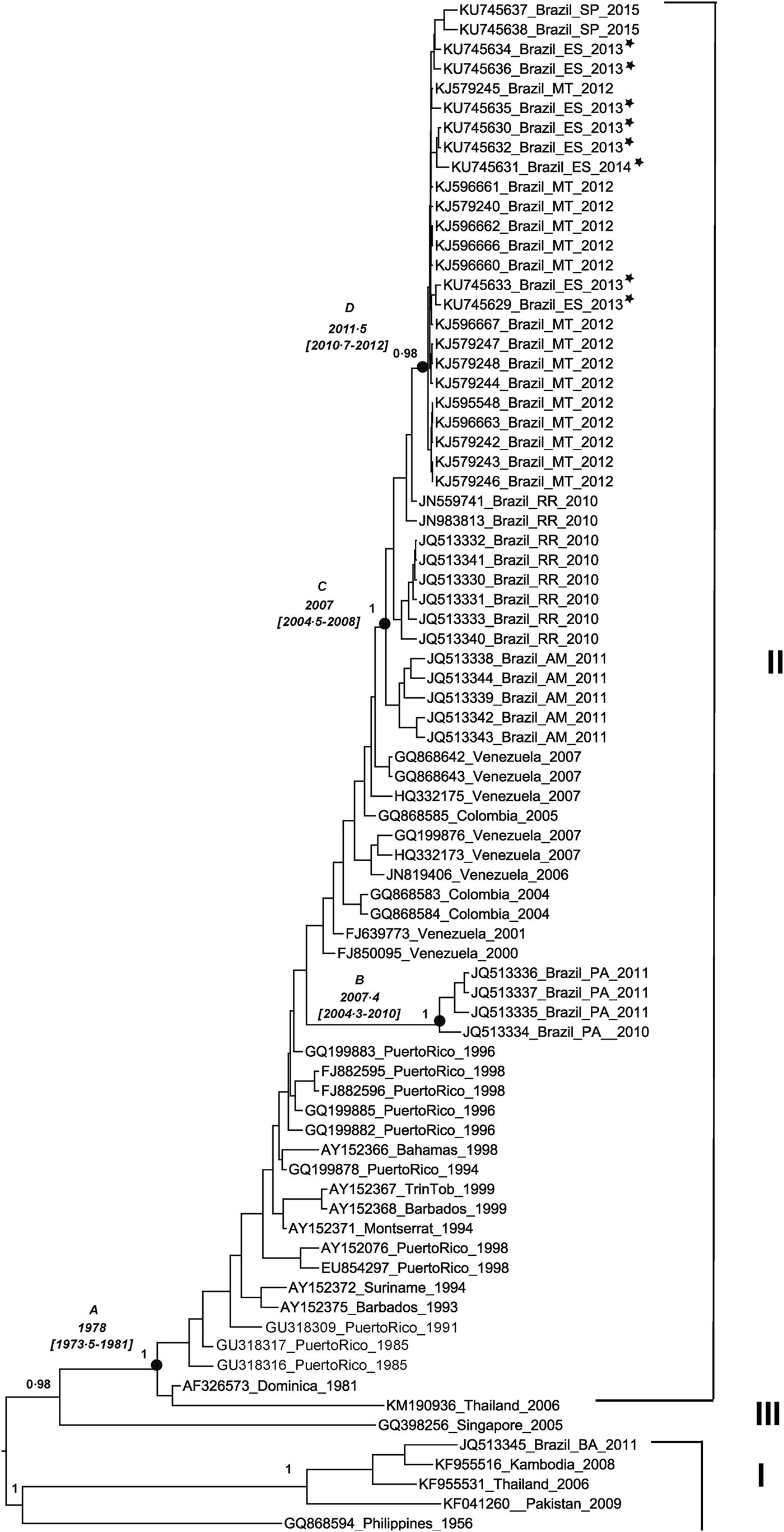
Fig. 1. Maximum clade credibility tree of dengue virus serotype 4. The phylogenetic tree shows the genotypes I, II and III of dengue virus serotype 4 (DENV-4). Partial envelope gene sequences of DENV-4 (1300 nucleotides) from Espírito Santo state (n = 8) and envelope gene sequences from GenBank were analysed. The phylogenetic tree shows the posterior probability values in the branches and the dates of the nodes (A, B, C and D), with lower and upper values within parenthesis. The length of the branches corresponds to sampling time. Sequence identifications include the GenBank code, the place and the year of the sample collection. The abbreviations in the sequence identifications correspond to the name of the following Brazilian states: Espírito Santo (ES), São Paulo (SP), Mato Grosso (MT), Roraima (RR), Amazonas (AM), Pará (PA) and Bahia (BA). The sequences from Espírito Santo state are marked with an asterisk.
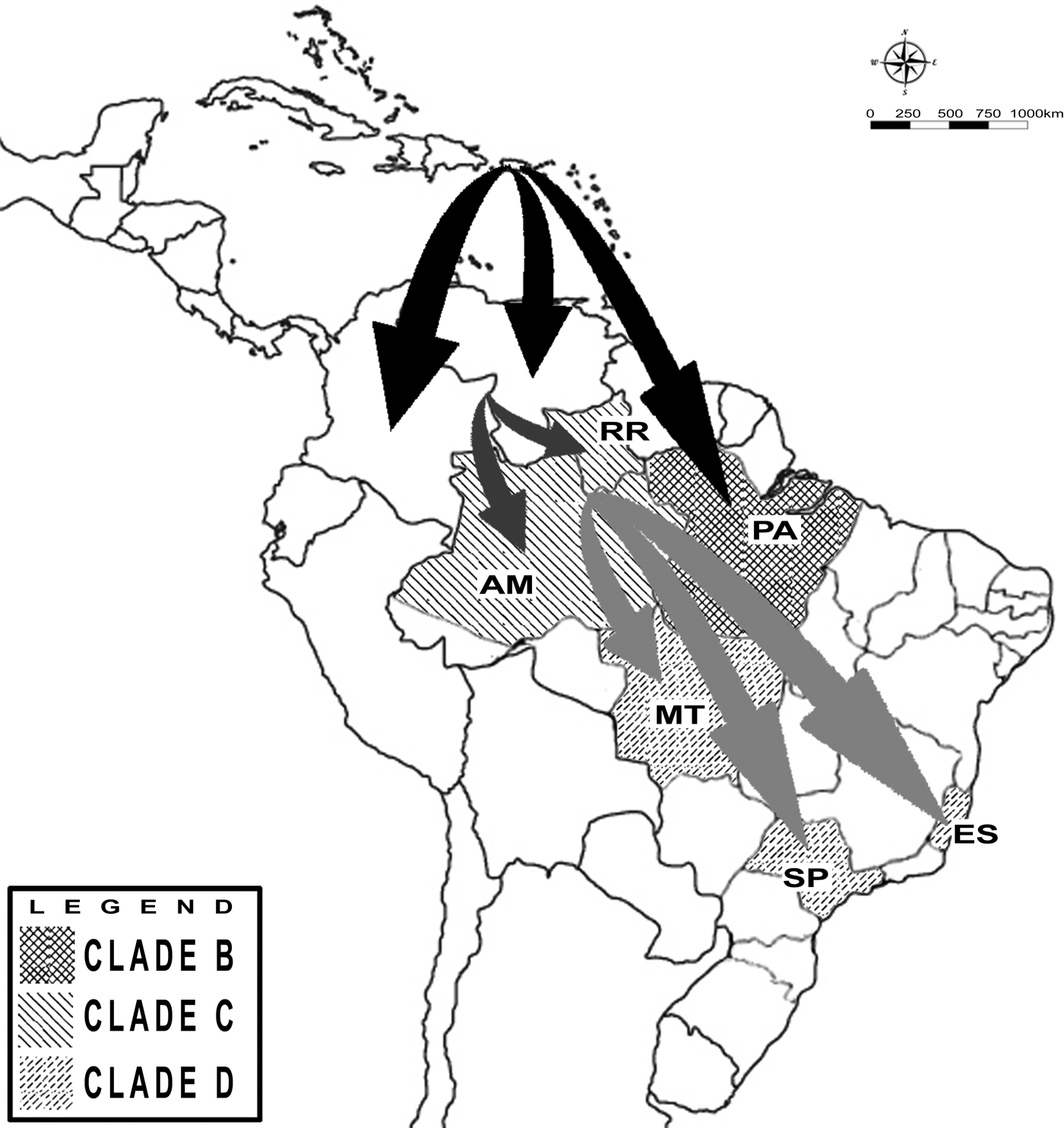
Fig. 2. Map showing the clades identified in the Brazilian states and possible routes for dengue introduction in South America and for dengue dispersion in Brazil. The abbreviations correspond to the name of the following Brazilian states: Espírito Santo (ES), São Paulo (SP), Mato Grosso (MT), Roraima (RR), Amazonas (AM), Pará (PA) and Bahia (BA).
The sequences obtained in this study were deposited in GenBank under the codes from KU745629 to KU745638.
DISCUSSION
The phylogenetic reconstruction based on a partial envelope gene sequence indicates the presence of the DENV-4 genotype II in Espírito Santo state in 2013 and 2014. The genotype II is the most prevalent DENV-4 genotype on the American continent [Reference Nunes19], where it was introduced between the 1970s and the beginning of the 1980s [Reference Nunes19–Reference Allicock21]. Puerto Rico is the possible origin of strains that spread to South America in the 1980s, corroborating the inferences on this country as a source of DENV-4 population in waves of dispersion for the continent [Reference Allicock21].
According to the present results, Brazil suffered at least two distinct introductions of DENV-4 genotype II in the 2000s, both in the northern region. One introduction, apparently restricted to Pará state, occurred between 2004 and 2010 by the direct importation of viruses from Puerto Rico, which remains an important route for dengue virus circulation between the Caribbean islands and South America. The second introduction was observed in the states of Amazonas and Roraima, with the probable importation of strains from Venezuela and Colombia, as anteriorly demonstrated [Reference Nunes19]. This introduction occurred between 2004 and 2008, at least 2 years before the first official report of a new DENV-4 circulation in Brazil, which occurred in Roraima in 2010. Thereby, DENV-4 circulated undetected for years, probably due to a restricted circulation in populations where other serotypes were already established [Reference Romano22, Reference Souza23]. This finding reinforces the importance of intensifying the viral surveillance in the Brazilian public health system, enabling the prediction of potential epidemics due to the circulation of a virus in an immunologically naive population. The present results also support the relevance of an integrated epidemiological surveillance in the Americas, since the identification of a new dengue virus strain in bordering countries is a predictor for its future introduction in surrounding territories, including foreign areas. Therefore, the close relationship of northern Brazilian states with Venezuela and Colombia may have contributed to the entry of humans infected by the virus into Brazil.
The strains of DENV-4 genotype II introduced in Roraima state are likely to have spread broadly in the country, generating a more recent lineage that emerged in Brazil around 2011 [Reference Souza23]. The strains of Espírito Santo state are grouped into this lineage. They are closely related to viruses from other Brazilian states, such as Mato Grosso and São Paulo, which implies that the DENV-4 genotype II found in Espírito Santo state was introduced from other Brazilian states, as similarly suggested previously for dengue virus serotype 2 [Reference Dettogni and Louro24]. Due to the absence of immunological protection against DENV-4 in the population of Espírito Santo state, the epidemic that followed the introduction and broad spread of this serotype, in 2013, accounted for the highest number of dengue fever cases reported to date in the state. These findings demonstrate a fast spread of dengue virus between Brazilian states, established by an internal route of intense viral circulation, which was able to reach distant territories, such as Espírito Santo state, located in the littoral of the southeast region, few years after the first report of DENV-4 in the northern area of Brazil. Espírito Santo state is located more than 3300 km far from Roraima, the first state to report DENV-4.
Despite the limited sample size, the present study was able to determine the relation of the DENV-4 circulating in Espírito Santo state with those from other regions for the first time. This information is relevant in order to understand the dynamic of viral circulation and to estimate how fast a new serotype can circulate, supporting the public health sector in the preparation to future viral introductions. The results also increase the knowledge about the most recent dengue virus serotype identified in the country, which in Espírito Santo state was the main agent involved in the most extensive epidemic recorded so far there.
The present study evidences the importance of considering the dynamic of dengue fever occurrence beyond local aspects. Since neighbouring countries and states are involved in the disease spread, the intensity of human movement across the territories might have facilitated the dispersion of the dengue virus among their populations. This factor must be considered in the public health evaluations about dengue fever.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the Health Department of Vitória municipality, for the logistic support in the collection of samples. This work was supported by the Coordination for the Improvement of Higher Education Personnel (CAPES) (C.R.V., grant number 9589-13-9); the São Paulo State Foundation (FAPESP) (C.M.R, grant number 2012/15381-7); and the Brazilian National Council for Scientific and Technological Development (CNPq) (C.C.J., grant number 482261/2010-2). The CIHLMU Centre for International Health at Medical Centre of the University of Munich is funded by the Federal Ministry for Economic Cooperation and Development (BMZ) and the German Academic Exchange Services (DAAD) through the Excellence Centres for Exchange and Development (EXCEED). The sponsors had no involvement in the study design, in the data collection, in the analysis and interpretation of data, in the writing of the manuscript, and in the decision to submit the article for publication.
DECLARATION OF INTEREST
None.





