Introduction
Phenacoccus solenopsis (P. solenopsis) Tinsley (Hemiptera: Sternorrhyncha: Coccoidea: Pseudococcidae) was first reported as a cotton pest in Texas, USA (Fuchs et al., Reference Fuchs, Stewart, Minzenmayer and Rose1991). Given the high variation of this species in terms of its morphological characteristics, biological adaptability, and ecological adjustability, it has reportedly settled in over 35 localities around the world, including Central America, the Caribbean, Ecuador (Ben-Dov, Reference Ben-Dov1994), Chile (Larrain, Reference Larrain2002), Argentina (Granara de Willink, Reference Granara de Willink2003), Brazil (Culik & Gullan, Reference Culik and Gullan2005), Pakistan, India (Karar, Reference Karar2008), Nigeria, Sri Lanka, and recently, China (Wang et al., Reference Wang, Wu and Zhang2009).
To adapt to a new environment, an invasive species may undergo a series of genetic and physiological changes. Several studies found that an invasive species can adapt through addictive genetic variation, epistasis, hybridization, genetic tradeoff, and a small number of gene and genomic rearrangements (Rhymer & Simberloff, Reference Rhymer and Simberloff1996; Lee, Reference Lee1999, Reference Lee2002; Tsutsui et al., Reference Tsutsui, Suarez, Holway and Case2000). In genetic variation, invasive species typically exhibit a broad range of behavior adaptability. For example, Capsella bursa-pastoris blooms earlier in the desert than on the beach or in snow forests (Neuffer & Hurka, Reference Neuffer and Hurka2002), and almost all invasive weeds have smaller seeds and shorter flowering times (Maillet & Lopez-Garcia, Reference Maillet and Lopez-Garcia2000).
Thus, the genetic or behavioral variation of a newly imported species somehow reflects its relationship with the population of the surrounding area. Genetic data provide information on the source of the invading population as well as hypotheses on the environmental and evolutionary factors that ensure the success of biological invasions (Sax et al., Reference Sax, Stachowicz and Gaines2005).
P. solenopsis, a notorious pest in cotton and a wide array of host plants, was first discovered in Guangzhou, China and has spread to several other provinces since then (Ma et al., Reference Ma, Hu, Liu and Peng2009). However, whether this insect has its own genetic and behavioral differences from native species remains unclear. Data illustrating whether relationships exist among the same-species populations from different areas after invasion are also unavailable. Thus, we obtained 31 P. solenopsis samples from 25 different locations in Southern China (fig. 1) and three samples from Pakistan to identify the relationship between the Chinese and Pakistani species. To our knowledge, the mitochondrial DNA (mtDNA) and internal transcribed spacer region of the nuclear ribosomal cistron (18S-5.8S-26S) show sufficiently high rates of nucleotide substitution (Hebert et al., Reference Hebert, Cywinska and Ball2003; Pons et al., Reference Pons, Barraclough, Theodorides, Cardoso and Vogler2004; Kress et al., Reference Kress, Wurdack, Zimmer, Weigt and Janzen2005; Havill et al., Reference Havill, Foottit and von Dohlen2007; Bellemain et al., Reference Bellemain, Carlsen, Brochmann, Coissac, Taberlet and Kauserud2010). These genes are often used to evaluate phylogenetic relationships among closely related species or genetically heterogeneous populations of a single species. In our research, partial sequences of the cytochrome oxidase subunit I (COI) gene were sequenced within the samples to infer the invasive status and variation information regarding P. solenopsis in China.

Fig. 1. Map showing the distribution of P. solenopsis Tinsley across Southern China. Thirty-one samples were collected from 25 different locations.
Materials and methods
Sampling collection
The P. solenopsis samples used in the present investigation were obtained from three provinces in Southern China, at the port, Pakistan (three samples), Vietnam (one sample), and the USA (two samples). Maconellicoccus hirsutus from Southern China was included as an outgroup. These samples were initially classified based on their morphology. Figure 1 gives an overview of the geographical distribution of the samples. Details regarding the sampling locations are provided in table 1. Live insects were carefully removed from the host plants. Once collected, the insects were immersed in 95% ethanol. Voucher samples were preserved at the Plant Quarantine Laboratory, Guangdong Inspection and Quarantine Technology Center.
Table 1. Information regarding the samples.

DNA extraction, amplification, and sequencing
All specimens were examined for the presence of parasitoids using a microscope. Total genomic DNA was extracted from the entire body of the P. solenopsis sample using the DNeasy Blood and Tissue Kit (QIAGEN) according to the manufacturer's instructions. DNA concentration and purity were estimated by means of absorbance at 260–280 nm in a NanoDrop ND-1000 (NanoDrop Technologies, Willminton, USA) spectrophotometer. Primers C1J2195 TTGATTYTTTGGTCATCCAGAAGT and TL2N3014 TCCAATGCACTAATCTGCCATATTA (Simon et al., Reference Simon, Frati, Beckenbach, Crespi, Liu and Flook1994) were initially used to amplify 840 bp of the mitochondrial COI gene. All samples were successfully amplified.
We amplified a partial sequence of the COI gene for phylogenetic analysis. The reaction was conducted in a thermal cycler in a 30 μl system composed of 3 μl of PCR buffer, 2.4 μl of dNTP mixture, 0.15 μl of Taq DNA polymerase, 1 μl of each primer, 1.8 μl of the DNA template, and ddH2O added to a final reaction volume of 30 μl. The PCRs were conducted in Eppendorf Mastercycler Thermal Cyclers using the following profile: an initial step of 4 min at 94 °C, followed by 35 cycles of 30 s at 95 °C, 45 s at 48 °C, and 1 min at 72 °C, which was followed by a final extension of 5 min at 72 °C. The PCR products were evaluated by placing 3 μl of the product on 1.2% agarose gel. DNA identity was then confirmed via sequencing using an ABI 3730xl sequencer (BGI Life Tech Co. Ltd., Shenzhen, China). The sequences of all haplotypes of the mealybug species were deposited in Gen Bank under accession Nos. KF878037 to KF878073 (COI) (table 1). The COI sequences of the P. solenopsis from India and the USA were obtained from Gen Bank with accession Nos. KC985429 to KC985430, JN112802, and EU267208 to EU267210.
Data analysis
We performed a BLAST search on NCBI (http://www.ncbi.nlm.nih.gov/) after obtaining the sequence of the PCR products. Once the high identity score (above 99%) of the P. solenopsis COI gene was obtained, we aligned the sequence data using ClustalW2 (Thompson et al., Reference Thompson, Higgins and Gibson1994) at the default parameter settings. Small changes were made to the alignment. The hyper-variable regions were excluded from further analysis because of the ambiguity of the alignment (Swofford et al., Reference Swofford, Olsen, Waddell, Hillis, Hillis, Moritz and Mable1996).
Unique haplotypes were identified using ARLEQUIN ver. 3.5. Descriptive statistics (number of variable sites, number of haplotypes, haplotype diversity, nucleotide diversity, and average number of nucleotide differences between haplotypes) were calculated using DNASP ver. 5.0.
An AMOVA hierarchical analysis of variance was performed using ARLEQUIN to partition the total variance in its components among groups, among populations, and within populations based on the groups inferred by the AMOVA analysis. The median joining (MJ) networks of the haplotypes of each of the COI genes was constructed using NETWORK ver. 4.6 to examine the evolutionary relationships among haplotypes.
A phylogenetic analysis of the COI based on the maximum likelihood (ML) method was performed separately using MEGA4 (Tamura et al., Reference Tamura, Dudley, Nei and Kumar2007) under the Kimura 2-parameter model and with the nearest-neighbor-interchange by means of the ML Heuristic method. The reliability of the branches was estimated using 1000 bootstraps.
Results
Amplification result and sequence information
We successfully amplified the COI in the samples and obtained bands of approximately 800 bp on the gel. We aligned the sequence once the PCR products were sequenced. Several base pairs were removed because of ambiguous alignment, which resulted in a final count of 852 COI base pairs. We determined 23 variable sites at the 84, 114, 145, 147, 168, 174, 189, 324, 354, 381, 399, 400, 402, 417, 442, 450, 598, 603, 636, 648, 660, 733, and 735 sites (table 2).
Table 2. Information regarding the base pair variations in the haplotypes.
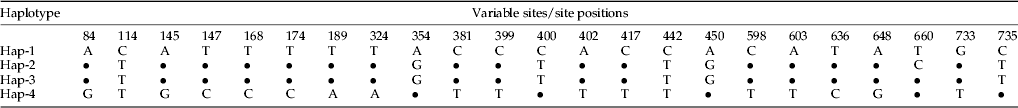
Population and haplotype division
The samples from Southern China were found to include two COI haplotypes characterized by 23 variable sites of COI. Two other haplotypes were observed among the samples from the USA, one of which was from California and another in Arizona. Most P. solenopsis populations in Southern China possessed haplotype 2. Several samples possessed haplotypes 2 and 3, including those from the Guangdong, Guangxi, and Hainan populations in Southern China, as well as from the Pakistan population, Hanoi population in Vietnam, and Delhi population in India (table 3). To determine the source of the introduction of P. solenopsis into China, the COI sequences we analyzed included not only all of the P. solenopsis samples we had obtained but also those from the USA and India, which were obtained by downloading homologous mtDNA sequences from Gen Bank.
Table 3. Geographic distribution frequencies of different haplotypes in populations of P. solenopsis.

Genetic diversity and distances
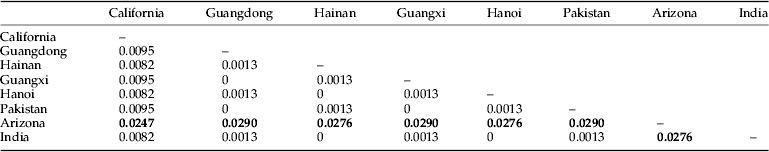
Haplotype 2 (populations from Guangdong, Guangxi, Hainan, and Pakistan) was the predominant haplotype in Southern China with a frequency of 0.535. Haplotypes 3 (populations in Guangdong, Guangxi, Hainan, Pakistan, Hanoi, and Delhi), 1 (population in California), and 4 (population in Arizona) had frequencies of 0.326, 0.070, and 0.070, respectively (table 3). The Pakistan population had the highest haplotype diversity (0.66667) and nucleotide diversity (0.00090) among all populations. By contrast, the haplotype and nucleotide diversities had no significant differences among all population from India, Vietnam, and the USA. In Southern China, the haplotype and nucleotide diversities were 0.39916 and 0.00054, respectively. The highest haplotype diversity was found in the Hainan population (0.53571) followed by the Guangdong population (0.43956), and the Guangxi population had the lowest haplotype diversity (0.22222). The average number of nucleotide differences was 3.885 for the species.
The genetic distances among the various populations of P. solenopsis varied from 0 to 0.0290 (table 4). No significant genetic differentiation was observed among the populations in Southern China (genetic distances ranged from 0 to 0.0095) as well as among the populations in India, Vietnam, and Pakistan. Conversely, significant genetic differentiation was detected from the population in Arizona.
Table 4. Genetic distances among various populations of P. solenopsis.
Genetic structure
The eight populations were clustered in terms of geographic location as follows: USA (California, Arizona); China (Guangdong, Guangxi, Hainan); South Asia (Pakistan, India); and Southeast Asia (Vietnam). These four geographically well-defined regions are discussed in the following presentation.
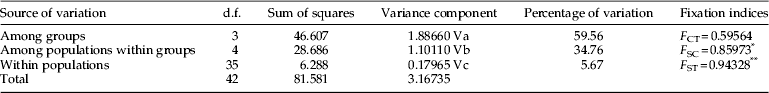
The AMOVA revealed that a portion of genetic differentiation was partitioned among groups (59.56% based on COI sequences) and within populations (5.67%), whereas excessive genetic differentiation was observed between populations within each of the four groups identified (34.76%) (table 5). Accordingly, differentiation was highly significant within populations (F ST) and among populations between groups (F SC).
Table 5. Partitioning of genetic variation at different hierarchical levels.
*P<0.05, **P<0.01.
MJ networks of haplotypes
MJ networks reconstructed from the haplotypes of the COI gene are shown in fig 2. Except for the USA samples, all samples from Southern China, Pakistan, Vietnam, and India had Hap-2 and Hap-3. Most samples from the Guangdong, Guangxi, and Hainan populations in the China group and two Pakistan samples in the South Asia group shared Hap-2. By contrast, a few samples from the Guangdong, Guangxi, and Hainan populations, one sample from the Vietnam-Hanoi population, one sample from the Pakistan population, and a few samples from the India-Delhi population shared Hap-3. The MJ networks of the COI haplotypes revealed that P. solenopsis has spread in Southern China.

Fig. 2. MJ networks of haplotypes. The pie area is proportional to haplotype frequency. Horizontal line: USA group; oblique line: China group; black: Southeast Asia group; grey: South Asia group; and white: inferred haplotypes.
Phylogenetic analysis and the potential distribution patterns of P. solenopsis in Southern China
We generated the ML trees using the aligned COI sequence. The tree included two distinct clades: one clade consisted of samples from the USA, and another clade consisted of samples from China, Vietnam, India, and Pakistan. However, slight differences among the Guangdong, Guangxi, and Hainan populations in the China group were observed (fig. 3). These data suggest that the invasion of P. solenopsis into China may have originated from Pakistan rather than from America. After their invasion into China, the P. solenopsis population may have undergone specific genetic-level variations to adapt to the new environment.

Fig. 3. Phylogenetic tree of partial COI sequences (852 bp) in the P. solenopsis regions obtained using the ML method. The numbers above the branches indicate bootstrap values (>50%, 1000 replicates). Maconellicoccus hirsutus was included as an outgroup based on the COI sequences.
Discussion
Variation of P. solenopsis in Southern China
Based on our knowledge, this study is the first to report on the molecular identification of P. solenopsis in Southern China based on the mtDNA COI gene. The P. solenopsis samples from the Guangdong, Guangxi, and Hainan populations in Southern China were found to include two COI haplotypes characterized by 23 variable sites of COI, which show limited genetic variation in the mealybugs in Southern China.
Although a number of structures were identified using AMOVA, which led to the partitioning of the eight populations being studied into four groups, namely, China, USA, South Asia, and Southeast Asia, the overall level of differentiation was low. Based on AMOVA results, a variability of more than 5% was observed within populations.
Based on the partial sequence variation of COI within the geographical P. solenopsis population in Southern China, we can infer that this species has undergone a series of genetic-level variations since it invaded China. To our knowledge, once an invasive species invades a new place, the main selective pressure originates from the change in environment (Sakai et al., Reference Sakai, Allendorf, Holt, Lodge, Molofsky, With, Baughman, Cabin, Cohen, Ellstrand and Mccauley2001). To adapt to this new environment, the invasive species must also change. Moreover, the invasive species can inherit this adaptation. P. solenopsis has spread to several other countries since 1991, and a previous research has reported that P. solenopsis underwent changes in a new environment (Karar, Reference Karar2008). Genetic variations in P. solenopsis may occur in a new habitat, and this variation may accelerate the invasion of P. solenopsis. Thus, questions of whether genetic-level variations in P. solenopsis in China and whether this variation affects the expansion of P. solenopsis still remain.
Possible source of P. solenopsis in Southern China
The phylogenetic tree showed that two divergent clades exist in this mealybug species. The major clade contained sequences of P. solenopsis from Guangdong, Guangxi, and Hainan in Southern China, Pakistan, Delhi in India, and Hanoi in Vietnam, whereas the other clade-contained sequences of P. solenopsis from the USA. The low genetic distance between the Guangdong and Guangxi populations from the China group and the Pakistan population in the South Asia group was 0, whereas the genetic distance between the Guangdong and Guangxi populations from the China group and the Arizona population from the USA group was 0.029. These data show that the P. solenopsis found in China may have originated from Pakistan rather than from America. These outcomes are in agreement with previous studies, wherein P. solenopsis was analyzed using the mitochondrial COI gene (Chu et al., Reference Chu, Liu and Fu2009; Chen et al., Reference Chen, Zhang, Fu, Xu, Deng and Zhang2012).
The genetic distance of P. solenopsis ranged from 0 to 0.029 when samples from the China, Pakistan, India, Vietnam, and USA populations were compared, and the genetic differentiation within populations in Southern China was low. Although P. solenopsis invaded China within 5 years, we can still infer that the P. solenopsis that invaded China were still in their primary stages.
The genetic data on which the source of the invading populations is based confirm the hypotheses concerning the environmental and evolutionary factors that ensure the success of biological invasions (Sax et al., Reference Sax, Stachowicz and Gaines2005). A recent study identified the global invasion of imported red fire ant using genetic data (Ascunce et al., Reference Ascunce, Yang, Oakey, Calcaterra, Wu, Shih, Goudet, Ross and Shoemaker2011). We aim to identify the potential source of P. solenopsis in China. Therefore, we compared the genetic data of P. solenopsis from Southern China, America, India, Vietnam, and Pakistan. We determined that all of the COI data samples from China had a closer relationship with those from Pakistan rather than those from America. These results suggest that the P. solenopsis in China may not have originated from the USA. More data are required to determine whether the P. solenopsis in China originated from Pakistan.
Spread of P. solenopsis in Southern China
Given the rapid development of international trade and tourism, P. solenopsis has been widely distributed worldwide. This species has caused severe economic losses over the last two decades in many countries. At present, many countries and regions where P. solenopsis are found include Mexico, USA, Cuba, Jamaica, Guatemala, Dominica, Ecuador (Ben-Dov, Reference Ben-Dov1994), Panama, Brazil, Chile (Larrain, Reference Larrain2002), Argentina (Granara de Willink, Reference Granara de Willink2003), Nigeria, Benin, Cameroon, New Caledonia, Pakistan, India (Karar, Reference Karar2008), and Thailand. In China, P. solenopsis was first found in 2008 in Guangzhou City (Ma et al., Reference Ma, Hu, Liu and Peng2009). Since its discovery, P. solenopsis has also been observed in Guangxi, Hainan, and other provinces (Wang et al., Reference Wang, Wu and Zhang2009). The MJ networks of COI haplotypes revealed that P. solenopsis has expanded in Southern China.
Based on the COI data, we determined that most of the P. solenopsis in Southern China, including those found within the same provinces and those in different provinces, belong to the same clade in the Pakistan tree of P. solenopsis. However, the samples obtained from the same provinces displayed slight differences with regard to COI sequence. This finding suggests that since P. solenopsis invaded China, this species may have spread from one place to another, which resulted in variations.
The data obtained in this study show that the source of the populations of P. solenopsis in Southern China might have originated from Pakistan instead of the USA. In future studies, we should collect more samples from all regions in China where P. solenopsis exist and use more types of markers to analyze the genetic differentiation and genetic structure of the species. These studies will help us understand the invasion history, invasion pattern, and source of this invasive population in Southern China. This molecular ecological evolution information on mealybugs may help us eliminate the invasion pathway or introduce biological control agents as well as learn from the measures taken by other countries regarding this matter.
Acknowledgements
We express our heartfelt thanks to the editor and two anonymous reviewers, for critical reading, insightful comments, and suggestions on this manuscript. This study was supported by the project of Science & Technology of General Administration of Quality Supervision, Inspection and Quarantine of the People's Republic of China (grant no. 2012IK271) and Science & Technology of Huizhou City, Guangdong Province, China (grant no. 2013B040009004) and National ‘Twelfth Five-Year’ Plan program for Science & Technology Support (grant no. 2012BAK11B01). The English writing of the manuscript was polished by EnPapers.











