Depressed inflammatory immune competence is characteristic of acute pre-pubescent protein–energy malnutrition in its most severe forms, i.e. marasmus, kwashiorkor and marasmic kwashiorkor, and is widely regarded as part of a generalised, unregulated immunological disintegration process(Reference Woodward, Suskind and Tontisirin1). However, this paradigm is under challenge by the Tolerance Model which proposes a tightly regulated, non-inflammatory form of immune competence that persists even in advanced weight loss, the presumptive benefit being a reduced risk of inflammatory reactions generated against catabolically released self-antigens(Reference Hillyer, Dao and Niemiec2–Reference Monk and Woodward4).
The Tolerance Model centres on three anti-inflammatory mediators, namely the glucocorticoids, transforming growth factor (TGF)-β and IL-10, a hormonal triad that, as discussed elsewhere(Reference Hillyer, Dao and Niemiec2–Reference Monk, Makinen and Shrum5), plays a determining role in physiological anti-inflammatory control and the maintenance of self-tolerance. Recently, high constitutive blood levels of each member of this anti-inflammatory triad have been reported to persist into the advanced stages of malnutrition in weanling mouse models of marasmus and incipient kwashiorkor(Reference Hillyer, Dao and Niemiec2–Reference Monk, Makinen and Shrum5). By contrast, the constitutive blood levels of the potently inflammatory cytokine interferon-γ are low in the same mouse models(Reference Hillyer, Maliwichi and Woodward6). Remarkably, the systemic rate of constitutive IL-10 production is at least sustained in the advanced stages of weight loss in these animal systems, and is even elevated in the marasmic condition(Reference Monk and Woodward3). The Tolerance Model predicts that malnutrition will reconfigure characteristically inflammatory lymphoid organs to exhibit a non-inflammatory nature while sustaining the capacities and character of non-inflammatory lymphoid sites. The objective of the present investigation was to test this prediction by determining whether acute forms of pre-pubescent malnutrition influence mRNA expression of genes critical to tolerogenic regulation in the spleen and small intestine, lymphoid organs that typically support inflammatory and non-inflammatory immune responses, respectively(Reference Westermann and Pabst7, Reference Mestecky, Russell and Elson8). In the present investigation, therefore, gene transcript expression served as a marker relating directly to synthetic potential with respect to the mediators selected for study.
Materials and methods
Animals and housing facilities
Male and female C57BL/6J mice were obtained from an in-house breeding colony. Caging and housing conditions have been described(Reference Hillyer, Dao and Niemiec2–Reference Hillyer, Maliwichi and Woodward6, Reference Woods and Woodward9). In particular, the mice were maintained in clean, conventional conditions that were not specific pathogen free. The present investigation was approved by the Animal Care Committee of the University of Guelph in accordance with the Canadian Council on Animal Care.
Experimental animals, dietary protocols and experimental design
Animals (18 d old) were weaned and acclimatised overnight to a complete purified diet described elsewhere(Reference Hillyer, Dao and Niemiec2–Reference Hillyer, Maliwichi and Woodward6, Reference Woods and Woodward9). At 19 d of age, animals were randomly assigned to one of three experimental groups (three males and three females per group), namely age-matched controls consuming the complete diet ad libitum, a restricted-intake group consuming the complete diet in restricted daily quantities as described(Reference Hillyer, Dao and Niemiec2–Reference Hillyer, Maliwichi and Woodward6) or a low-protein group consuming ad libitum a 0·6 % crude protein diet (as fed) that was isoenergetic with the complete formulation which typically provides (as fed) 18 % crude protein(Reference Hillyer, Dao and Niemiec2–Reference Hillyer, Maliwichi and Woodward6, Reference Woods and Woodward9). The experimental period was 14 d, and animals were killed using CO2 followed by cervical dislocation without recovering consciousness.
RNA isolation
The spleen and small intestine were recovered aseptically and immediately immersed, intact, in ice-cold TriPure Isolation Reagent (‘TriPure’; Roche Diagnostics Corporation, Indianapolis, IN, USA). The spleen was immersed in 1 ml TriPure. The small intestine, excised proximally at the pyloric sphincter and distally at the ileocaecal junction, was immersed in 3 ml TriPure and its lumen was flushed with 10 ml of sterile 0·01 m-PBS (pH 7·3) containing 0·25 mm-EDTA followed by 10 ml TriPure. Subsequently, the tissues were immediately homogenised and centrifuged at 12 000 g for 10 min at 4°C. Total RNA was isolated from the resulting supernatant utilising the RNeasy Mini Kit (Qiagen, Inc., Mississauga, ON, Canada) according to the manufacturer's instructions and samples were treated with DNase to remove any DNA contamination.
Assessment of mRNA transcript levels
Reverse transcription of 1·5 μg of total RNA was achieved using Moloney Murine Leukemia Virus RT (Invitrogen, Carlsbad, CA, USA). Real-time RT-PCR was used to quantify mRNA expression and amplification was performed using the Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) and the 7900-HT Real-Time PCR System (Applied Biosystems). The primer sets for each cytokine, previously used successfully(Reference Barr, Brown and Ryan10), were as follows: IL-10 – 5′-ggttgccaagccttatcgga-3′ (forward) and 5′-acctgctccactgccttgct-3′ (reverse); IL-12p40 – 5′-ggaagcacggcagcagaata-3′ (forward) and 5′-aacttgagggagaagtaggaatgg-3′ (reverse); TGF-β1 – 5′-cacctgcaagaccatcgaca-3′ (forward) and 5′-cacgcgggtgacctctttag-3′ (reverse); forkhead box P3 (Foxp3) – 5′-ttggccagcgccatctt-3′ (forward) and 5′-tgcctcctccagagagaagtg-3′ (reverse). Relative gene expression was determined using the 2− ΔΔC T method(Reference Livak and Schmittgen11) with standardisation against β-actin for which the primer sets were 5′-tggaaatcctgtggcatccagtaaac-3′ (forward) and 5′-taaaacgcagctcagtaacagtccg-3′ (reverse).
Carcass composition
Animal carcasses were stored at − 20°C to await analysis of DM and total lipid content as described(Reference Hillyer, Dao and Niemiec2–Reference Hillyer, Maliwichi and Woodward6, Reference Woods and Woodward9).
Statistical analysis
The predetermined upper limit of probability for statistical significance throughout the present investigation was P ≤ 0·05, and analyses were conducted using the SAS system (SAS Institute, Cary, NC, USA) for Windows (version 9.0). Data were subjected to ANOVA followed, if justified, by Tukey's studentised range test. Datasets not exhibiting a normal distribution were transformed. Where transformation attempts failed, data were subjected to the Kruskal–Wallis test (χ2 approximation) followed, if justified by the statistical probability outcome (P ≤ 0·05), by Wilcoxon's two-sample test.
Results
Growth indices and food intakes
Performance indices are shown in Table 1. Food intakes and gains in carcass lean and fat tissue by the age-matched control group were comparable with previous outcomes(Reference Hillyer, Dao and Niemiec2–Reference Hillyer, Maliwichi and Woodward6, Reference Woods and Woodward9). Likewise, the malnourished groups exhibited characteristics similar to those reported previously in studies demonstrating reduced inflammatory immune competence in the same experimental systems(Reference Woods and Woodward9).
Table 1 Initial and final body weights, food intakes and carcass compositions
(Mean values with their standard errors)
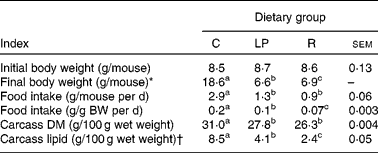
C, group that consumed the complete diet ad libitum; LP, group that consumed ad libitum an isoenergetic low-protein diet; R, group that consumed the complete diet in restricted daily quantities; BW, body weight.
a,b,c Mean values within a row with unlike superscript letters were significantly different (P ≤ 0·05; according to Tukey's studentised range test, unless a different statistical procedure is indicated).
* Kruskal–Wallis test of Wilcoxon rank sums that were as follows: C = 93·0; LP = 26·0; R = 52·0.
† From ANOVA of natural log-transformed data. Mean values are antilogs of mean values from log-transformed data.
Expression of transcripts for IL-10, transforming growth factor-β1 and IL-12p40
The quantity of mRNA detected for IL-10, TGF-β1 and IL-12p40 is expressed relative to the quantity of mRNA for β-actin in Fig. 1(a)–(c), respectively. Neither form of malnutrition influenced the mRNA transcript levels of the three cytokines in the small intestine, but several influences of malnutrition emerged in the spleen. First, splenic cytokine mRNA levels generally exceeded those in the small intestine. Moreover, the quantity of IL-10 mRNA was greater in the spleen of both malnourished groups than in the spleen of the age-matched control group, medians being approximately 2- and 5-fold higher in the low-protein and restricted-intake groups, respectively. Similarly, the median quantity of mRNA for TGF-β1 was 2-fold higher in the spleen of the restricted-intake group than in the spleen of the age-matched controls, and the quantity of this mRNA species was sustained in the spleen of the group consuming the low-protein diet. By contrast, the median quantity of splenic mRNA for IL-12p40 in the low-protein and restricted-intake groups was reduced by approximately 50 and 90 %, respectively, compared with the quantity in the spleen of the age-matched control group.

Fig. 1 Cytokine mRNA expression in lymphoid organs (small intestine, ■; spleen, ![]() ) assessed by real-time RT-PCR. C57BL/6J mice initially 19 d old were fed a complete purified diet ad libitum (age-matched controls, C), or were given the complete diet in restricted daily quantities (restricted-intake group, R), or were given free access to an isoenergetic low-protein diet containing 0·6 % crude protein (low-protein group, LP) for 14 d (six animals per dietary group). Total RNA was isolated from the spleen and small intestine. Relative abundance of (a) IL-10, (b) transforming growth factor (TGF)-β1, (c) IL-12p40 and (d) forkhead box P3 (Foxp3) mRNA was determined using the 2− ΔΔCT method with standardisation against β-actin. Bars represent medians and the data were analysed by the Kruskal–Wallis test (P < 0·05), followed by Wilcoxon's two-sample test. The Wilcoxon rank sums within each lymphoid site (C, LP and R, respectively) were as follows: IL-10 – 59, 29, 87 (intestine) and 131, 164, 196 (spleen); TGF-β1 – 72, 52, 48 (intestine) and 141, 161, 192 (spleen); IL-12p40 – 58·5, 70, 83 (intestine) and 195, 158·5, 101 (spleen); Foxp3 – 76, 89, 120 (intestine) and 264, 275, 275 (spleen). For each gene of interest, within anatomical sites, bars not sharing an upper-case letter denote differences between dietary groups (P ≤ 0·05) wherein letter assignments are as follows: intestine (A,B) and spleen (X,Y,Z). Statistical probability values cited in the figure pertain to comparisons between anatomical sites within dietary groups.
) assessed by real-time RT-PCR. C57BL/6J mice initially 19 d old were fed a complete purified diet ad libitum (age-matched controls, C), or were given the complete diet in restricted daily quantities (restricted-intake group, R), or were given free access to an isoenergetic low-protein diet containing 0·6 % crude protein (low-protein group, LP) for 14 d (six animals per dietary group). Total RNA was isolated from the spleen and small intestine. Relative abundance of (a) IL-10, (b) transforming growth factor (TGF)-β1, (c) IL-12p40 and (d) forkhead box P3 (Foxp3) mRNA was determined using the 2− ΔΔCT method with standardisation against β-actin. Bars represent medians and the data were analysed by the Kruskal–Wallis test (P < 0·05), followed by Wilcoxon's two-sample test. The Wilcoxon rank sums within each lymphoid site (C, LP and R, respectively) were as follows: IL-10 – 59, 29, 87 (intestine) and 131, 164, 196 (spleen); TGF-β1 – 72, 52, 48 (intestine) and 141, 161, 192 (spleen); IL-12p40 – 58·5, 70, 83 (intestine) and 195, 158·5, 101 (spleen); Foxp3 – 76, 89, 120 (intestine) and 264, 275, 275 (spleen). For each gene of interest, within anatomical sites, bars not sharing an upper-case letter denote differences between dietary groups (P ≤ 0·05) wherein letter assignments are as follows: intestine (A,B) and spleen (X,Y,Z). Statistical probability values cited in the figure pertain to comparisons between anatomical sites within dietary groups.
Expression of the transcript for forkhead box P3
The quantity of Foxp3 mRNA, expressed relative to mRNA for β-actin, is shown in Fig. 1(d) and was higher in the spleen than in the small intestine regardless of the diet. Neither form of malnutrition influenced the level of mRNA for Foxp3 in either the spleen or the small intestine.
Discussion
The present investigation centred on key inflammatory regulators in metabolically distinct murine models of acute pre-pubescent protein and energy deficit. IL-10 and TGF-β were selected as the two main soluble mediators of self-tolerance(Reference Li and Flavell12), whereas IL-12 was selected as a key inflammatory mediator(Reference Gutcher and Becher13, Reference Goriely, Neurath and Goldman14). Foxp3 was included to provide an index of tolerogenic cellular immune competence(Reference Sakaguchi, Ono and Setoguchi15). On the basis of these indices, acute malnutrition appeared to sustain the non-inflammatory nature of a mucosal immune organ, the intestine, while supporting an immunological reconfiguration towards a non-inflammatory and tolerogenic character within the spleen. Importantly, these findings emerged in both the restricted-intake model and the low-protein model, metabolically distinct forms of malnutrition that consistently reproduce the critical features of paediatric marasmus and incipient paediatric kwashiorkor, respectively(Reference Hillyer, Dao and Niemiec2–Reference Hillyer, Maliwichi and Woodward6, Reference Woods and Woodward9). The strategy of examining whole organs as the basic immunological unit (rather than isolating individual cell types) was essential to the objective of the study both because of the diverse splenic and intestinal cells that produce cytokines such as IL-10 and TGF-β(Reference Li and Flavell12) and because of the need to minimise procedurally related changes in gene transcript levels. The present investigation provides the first support for the Tolerance Model at the level of mRNA transcript expression.
The Tolerance Model predicts that secondary lymphoid organs designed for inflammatory responses will be reconfigured towards a non-inflammatory character by acute pre-pubescent malnutrition, whereas sites already providing a non-inflammatory type of defence will be sustained as such. The spleen was selected for the present investigation as the largest mammalian inflammatory lymphoid organ, whereas the intestine was included as the largest strictly non-inflammatory immunological organ(Reference Westermann and Pabst7). Consistent with the prediction of the Tolerance Model, the spleen in both models of malnutrition shifted dramatically towards a non-inflammatory cytokine-producing potential while the intestine, specifically designed to provide a non-inflammatory type of defence(Reference Mestecky, Russell and Elson8), was sustained in this distinctive character. In addition, the findings point to the spleen as a significant contributor to the high tissue fluid levels of IL-10 and TGF-β1(Reference Hillyer, Dao and Niemiec2–Reference Monk and Woodward4) and the high systemic IL-10 production rate(Reference Monk and Woodward3) reported in the same weanling models of acute malnutrition. Finally, it should be noted that the IL-12p40 subunit, pursued herein, serves as a marker for the inflammatory IL-12 dimer(Reference Gutcher and Becher13), and the balance between IL-12 and IL-10 exerts a controlling influence over T-cell-mediated inflammation(Reference Goriely, Neurath and Goldman14). Thus, a reduced expression of splenic IL-12 is critical to discernment of the immunological character of the forms of malnutrition examined herein. Remarkably, splenic expression of mRNA for this inflammatory cytokine was reduced in the marasmic model to a level indistinguishable from its expression in the intestine, a prototypical non-inflammatory lymphoid organ.
The systemic rate of constitutive IL-10 synthesis is sustained, or even increased, in the weanling models of acute malnutrition used herein(Reference Monk and Woodward3). By contrast, no information is available quantifying the rate at which TGF-β1 is synthesised in acute malnutrition, although very high blood levels of the biologically active cytokine are maintained(Reference Hillyer, Dao and Niemiec2, Reference Monk and Woodward4), as is reported also for IL-10(Reference Hillyer, Dao and Niemiec2, Reference Monk and Woodward3). The relationship between TGF-β1 mRNA and protein levels is complex and indirect because a large pool of this cytokine is bound to the extracellular matrix awaiting proteolytic release and activation(Reference Annes, Munger and Rifkin16). Nevertheless, the high tissue fluid levels of both TGF-β1 and IL-10 reported in the malnutrition systems of this investigation(Reference Hillyer, Dao and Niemiec2–Reference Monk and Woodward4) are reasonably attributable, at least partly, to a high availability of corresponding mRNA transcripts and, thus, to an increase in the synthesis of these tolerogenic cytokines.
The outcome pertaining to Foxp3 transcripts complements the findings regarding cytokine transcript levels in both models of acute malnutrition. Foxp3 is a transcription factor specifically identifying major subsets of anti-inflammatory, regulatory T-cells(Reference Sakaguchi, Ono and Setoguchi15). In view of the disproportionate involution of the T-cell compartment that is characteristic of acute malnutrition(Reference Woodward, Suskind and Tontisirin1), maintenance of Foxp3 transcript expression into advanced weight loss is remarkable and extends the present investigation to include tolerogenic cellular elements.
Despite limited supplies of energy and nitrogen, the potential for sustaining production of non-inflammatory mediators increased in both models of acute malnutrition and only the potential for synthesising an inflammatory mediator declined. In addition, the spleen emerges reconfigured and primed to function as a tolerogenic regulator in the malnourished. Collectively, these findings are incompatible with the model of chaotic immunological disintegration to which malnutrition-associated depression in inflammatory capacities is widely attributed. The Tolerance Model, however, now with support based on the expression of critical mRNA transcripts, comfortably predicts and accommodates the findings as part of a tightly regulated process aimed at establishing a non-inflammatory form of immune competence systemically in acute malnutrition.
Acknowledgements
This study was supported by a Postgraduate Scholarship and a Discovery Grant awarded, respectively, to J. M. M. and B. W. by NSERC of Canada. The authors gratefully acknowledge Ms Katazyna Stepien in connection with carcass analyses. J. M. M. and B. W. designed the study and wrote the manuscript. J. M. M. conducted the experimental work. J. M. M. and C. L. R. analysed the data. No relevant conflicts of interest arise.




