Schizophrenia (OMIM (Online Mendelian Inheritance in Man®): 181500) is a chronic, severe, and debilitating mental illness that generally presents in early adult life and is characterized by a disruption of perception and thinking. The lifetime prevalence of this disease is ~1% with little variation in prevalence seen throughout the world (Public Health Agency of Canada Steering Committee on Health — Adjusted Life Expectancy, 2013). The disease clusters in some families and has a high heritability estimate (80%; Sullivan et al., Reference Sullivan, Kendler and Neale2003). In fact, the best predictor of the occurrence of this disease is family history. The inheritance pattern of schizophrenia is complex. This complexity is reflected in the observation that monozygotic twins, who are said to share 100%, and dizygotic twins who are said to share 50%, of their genetic makeup are concordant in only 48% and 17% of cases, respectively (McGuffin et al., Reference McGuffin, Asherson, Owen and Farmer1994). These observations suggest a role for non-genetic and random genetic factors (O'Reilly & Singh, Reference O'Reilly and Singh1996; Singh & O'Reilly, Reference Singh and O'Reilly2009), including random developmental events (Singh et al., Reference Singh, McDonald, Murphy and O'Reilly2004), epigenetic mechanisms (Singh et al., Reference Singh, Murphy and O'Reilly2002), and environmental factors (Torrey et al., Reference Torrey, Miller, Rawlings and Yolken1997). Over 30 years of genetic research using linkage and association analysis have identified a number of promising linkages (Sullivan, Reference Sullivan2005) and candidate genes (Hamilton, Reference Hamilton2008; Karayiorgou & Gogos, Reference Karayiorgou and Gogos2006). Most of these results have been difficult to reliably replicate except in the case of a few variants, which have been associated across multiple studies, typically when large sample sizes are employed (Ripke et al., Reference Ripke, O'Dushlaine, Chambert, Moran, Kahler, Akterin and Sullivan2013; Torkamani et al., Reference Torkamani, Dean, Schork and Thomas2010). This difficulty in identifying causal genes for schizophrenia has been attributed to extensive heterogeneity, including different patients from the same family (Beckmann & Franzek, Reference Beckmann and Franzek2000). Application of genome-wide expression arrays in schizophrenia has identified a long list of genes with altered expression in the brain (McInnes & Lauriat, Reference McInnes and Lauriat2006; Verveer et al., Reference Verveer, Huizer, Fekkes and van Beveren2007) and blood tissue (Gladkevich et al., Reference Gladkevich, Kauffman and Korf2004; Tsuang et al., Reference Tsuang, Nossova, Yager, Tsuang, Guo, Shyu and Liew2005). However, altered expression of these genes cannot always be replicated and may be a secondary effect.
Recent advances in human genomics have helped in the identification of structural variants, termed copy number variants (CNVs), and opened a new direction in schizophrenia genetics research (Kirov, Reference Kirov2010). Copy number variants lead to deletions and duplications of a given segment of the genome. They are common (Conrad et al., Reference Conrad, Bird, Blackburne, Lindsay, Mamanova, Lee and Hurles2010) and widespread in the human genome (Iafrate et al., Reference Iafrate, Feuk, Rivera, Listewnik, Donahoe, Qi and Lee2004). By virtue of their variable size, they may directly disrupt multiple genes that are co-located (Feuk et al., Reference Feuk, Marshall, Wintle and Scherer2006). In addition to having a direct effect on the expression of the amplified or deleted genes (Stranger et al., Reference Stranger, Forrest, Dunning, Ingle, Beazley, Thorne and Dermitzakis2007), they may have indirect effects on gene expression extending upstream and downstream of the CNV region (Henrichsen et al., Reference Henrichsen, Vinckenbosch, Zollner, Chaignat, Pradervand, Schutz and Reymond2009). While most CNVs are polymorphic, some are generated de novo (Zogopoulos et al., Reference Zogopoulos, Ha, Naqib, Moore, Kim, Montpetit and Gallinger2007). The common CNVs in humans are believed to play a role in evolution (Lee & Scherer, Reference Lee and Scherer2010). They also underlie a significant proportion of variation in humans, including differences in cognitive, behavioral, and psychological features (Lee & Lupski, Reference Lee and Lupski2006). Further, they have been implicated across a wide variety of common disorders (Buchanan & Scherer, Reference Buchanan and Scherer2008; Stankiewicz & Lupski, Reference Stankiewicz and Lupski2010; Wellcome Trust Case Control Consortium et al., Reference Craddock, Hurles, Cardin, Pearson, Plagnol and Donnelly2010), including mental disorders (Feuk et al., Reference Feuk, Marshall, Wintle and Scherer2006; Lee & Lupski, Reference Lee and Lupski2006; McCarroll & Altshuler, Reference McCarroll and Altshuler2007), particularly autism (OMIM: 209850; Autism Genome Project Consortium et al., Reference Szatmari, Paterson, Zwaigenbaum, Roberts, Brian and Meyer2007; Glessner et al., Reference Glessner, Wang, Cai, Korvatska, Kim, Wood and Hakonarson2009; Moessner et al., Reference Moessner, Marshall, Sutcliffe, Skaug, Pinto, Vincent and Scherer2007; Sebat et al., Reference Sebat, Lakshmi, Malhotra, Troge, Lese-Martin, Walsh and Wigler2007; Wang et al., Reference Wang, Zhang, Ma, Bucan, Glessner, Abrahams and Hakonarson2009) and schizophrenia (Glessner et al., Reference Glessner, Reilly, Kim, Takahashi, Albano, Hou and Hakonarson2010; Kirov et al., Reference Kirov, Gumus, Chen, Norton, Georgieva, Sari and Ullmann2008; Need et al., Reference Need, Ge, Weale, Maia, Feng, Heinzen and Goldstein2009; Stefansson et al., Reference Stefansson, Rujescu, Cichon, Pietilainen, Ingason, Steinberg and Stefansson2008; Walsh et al., Reference Walsh, McClellan, McCarthy, Addington, Pierce, Cooper and Sebat2008; Xu et al., Reference Xu, Roos, Levy, van Rensburg, Gogos and Karayiorgou2008). The results, generated with increasing genomic coverage and numbers of patients, have identified a set of candidate CNVs. These include rare deletions at 1q21.1, 15q13.3, 15q11.2, and 22q11.2, as well as duplications at 16p11.2, 16p13.1, and 7q36.3 (Kirov et al., Reference Kirov, Gumus, Chen, Norton, Georgieva, Sari and Ullmann2008). In addition, various gene regions have been associated with copy number variation in schizophrenia, namely deletions of NRXN1 (Entrez Gene: 9378), APBA2 (Entrez Gene: 321), and CNTNAP2 (Entrez Gene: 26047; Friedman et al., Reference Friedman, Vrijenhoek, Markx, Janssen, van der Vliet, Faas and Veltman2008; Liu et al., Reference Liu, Abecasis, Heath, Knowles, Demars, Chen and Karayiorgou2002).
The findings in the field also suggest that, with few exceptions, schizophrenia is caused by aberrations in a relatively large number of genes, most with relatively small effects, that cumulatively produce a genetic predisposition. Some of these aberrations may be inherited while others may represent de novo events (Singh et al., Reference Singh, Castellani and O'Reilly2009). The field is starting to recognize that rare variants likely play a role in the causation of schizophrenia. This model is not compatible with traditional experiments in which a group of patients are compared with an equally large group of unaffected controls. In such an approach, adding more patients will add additional genetic heterogeneity across cases. This complexity is likely better approached by the precise genetic matching of patients with unaffected controls that can be achieved using monozygotic twins. Even if rare variants identified using this approach are limited to a given set of twins or a given family, they are likely to help in identifying the underlying pathways and genes involved in this disorder.
In this research, we used six pairs of monozygotic twins discordant (MZD) for schizophrenia and assessed the CNV differences between twin pairs. The resulting CNV differences are of interest in identifying patient-specific differences, including gene dosage changes that may differ in a MZD pair. Previous studies utilizing monozygotic twins have associated CNV and methylation differences between twins with various diseases. Using monozygotic twins, three somatic CNV events were found to be associated with discordance for congenital heart defects (Breckpot et al., Reference Breckpot, Thienpont, Gewillig, Allegaert, Vermeesch and Devriendt2012). Similarly, two de novo CNVs — a pre-twinning duplication and a post-twinning deletion were found to be associated with attention problems (Ehli et al., Reference Ehli, Abdellaoui, Hu, Hottenga, Kattenberg, van Beijsterveldt and Davies2012). Another study looking at Rett syndrome in discordant monozygotic twins found differences in deoxyribonucleic acid (DNA) methylation between twins detected in fibroblasts in the upstream region of genes involved in brain function to be associated with the disease (Miyake et al., Reference Miyake, Yang, Minakuchi, Ohori, Soutome, Hirasawa and Kubota2013). The results are twin-specific and trends are not always consistent (Bloom et al., Reference Bloom, Kahler, Collins, Chen, Cannon, Hultman and Sullivan2013; Halder et al., Reference Halder, Jain, Chaudhary and Varma2012; Maiti et al., Reference Maiti, Kumar, Castellani, O'Reilly and Singh2011). Some studies call CNV differences (Maiti et al., Reference Maiti, Kumar, Castellani, O'Reilly and Singh2011) while others call no difference in CNVs between MZ twins discordant for schizophrenia (Bloom et al., Reference Bloom, Kahler, Collins, Chen, Cannon, Hultman and Sullivan2013). In either case, the MZD strategy is effective in the identification of previously undiscovered genes in schizophrenia, particularly when combined with the use of multiple software programs. Given the high heterogeneity of this disorder, we would a priori expect many aberrations to be patient-specific. These patient-specific genetic changes can be best identified using nature's best match for each patient — their monozygotic twin. We have hypothesized that the discordance of monozygotic twins for schizophrenia may involve de novo mutations (DNM; Singh et al., Reference Singh, Castellani and O'Reilly2009). If that is so, we should be able to identify differences between MZD twins for schizophrenia that are de novo in nature and do not apply to all twin pairs, but instead show twin-pair specificity. In this report, we have employed a stringent copy number variation detection protocol using multiple CNV calling methods, and identified CNV differences between MZD for schizophrenia. The results support the potential presence of de novo CNVs that are compatible with the development of schizophrenia.
Materials and Methods
Ethics Statement and Clinical Background
This study received ethics approval by the University of Western Ontario's Committee on research involving human subjects. All subjects provided written informed consent to participate in this study and were interviewed by a psychiatrist (ROR) using the SCID-I (Structured Clinical Interview for DSM-IV Axis I Disorders; First et al., Reference First, Spitzer, Gibbon and Williams1996) and the SCID-II (Structured Clinical Interview for DSM-IV Axis II Disorders; First et al., Reference First, Gibbon, Spitzer, Williams and Benjamin1997). All of the patients were adults at the time of consent. Past clinical notes were obtained to aid diagnosis. Whole blood samples were obtained from each individual. The twin pairs studied ranged in the age from 20 to 53 years at the time of sample collection. Three of the pairs were female and three of the pairs were male. The twins were discordant for schizophrenia (defined as the time of first contact with mental health services because of symptoms of mental illness) for 4 to 31 years. The strategy used to generate and interpret the molecular results is outlined in the flowchart (Figure 1).
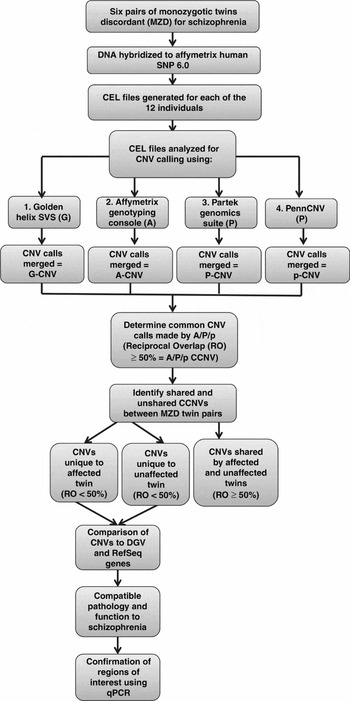
FIGURE 1 Flowchart of experimental design CCNV: Consistent Copy Number Variant.
DNA Preparation, Hybridization and CEL File Analysis
Deoxyribonucleic acid was extracted from whole blood using the PerfectPure DNA Blood Kit (http://www.5prime.com) following the manufacturer's protocol. Whole genome microarray analysis using the Affymetrix® Genome-Wide Human SNP Array 6.0™ was performed at the London Regional Genomics Center (LRGC) following the manufacturer's protocol. For downstream analysis of .cel files, Affymetrix® Genotyping Console 4.1.1™ (A), Partek® Genomics Suite™(P), PennCNV (p), and Golden Helix® SVS Suite 7.0™ (G) were used.
Calling and Merging of CNVs for Individual Genomes
The Hap Map 270 6.0 Array reference was utilized as a model reference file. Variants were identified as those DNA regions, which were called as copy number state 0, 1, 3, or 4+ by 10 or more consecutive markers on the chip. Recent literature suggests that a baseline of at least seven consecutive probes is necessary for reliable CNV detection (Wineinger & Tiwari, Reference Wineinger and Tiwari2012). In addition, only variants that were greater than 1 kb in size were classified as CNVs for the purposes of this study and only those identified by at least three software programs in the same individual were included in subsequent analysis. We used quantile normalization across all four software programs. Overlapping genes were identified using the UCSC (University of California, Santa Cruz) genome browser table view (NCBI36/hg18). Identification of CNVs was followed by merging of CNV calls within software programs and comparison of calls between software programs to identify CNVs identified by three or more software programs. We identified CNVs within each of the four software programs that were likely to be the same event using the following criteria: (1) CNVs had to be adjacent on the same chromosome (no other CNV call between them); (2) had to share the same gain/loss status; (3) adjacent calls were merged together into one single call, using gap ≤20% of total length. That is, if there are three genomic segments, A, B, and C, where A and C are both losses, we divided the length of gap B by the length of A + B + C, and if this fraction is ≤20%, then we merged A + B + C as a single CNV call.
Identification of Common CNV Calls for Individual Genomes
We utilized a reciprocal overlap (RO) formula. Copy number variants that shared 50% or more similarity with one another were classified as common. This is consistent with the definition of an overlapping CNV identified by other groups (Pang et al., Reference Pang, MacDonald, Pinto, Wei, Rafiq, Conrad and Scherer2010; Wain et al., Reference Wain, Pedroso, Landers, Breen, Shaw, Leigh and Al-Chalabi2009; Yavas et al., Reference Yavas, Koyuturk, Ozsoyoglu, Gould and LaFramboise2009). In other words, if at least half of the first CNV overlapped with the second CNV and vice versa they were considered to be the same event.
Comparison of CNV Calls Within Monozygotic Twin Pairs
The same RO definition was used to compare calls between monozygotic twin pairs. Copy number variants were compared between affected and unaffected twin pairs to determine which CNVs were shared and unshared between twins. Unshared CNVs between twins were then annotated with gene information and compared to CNVs in the Database of Genomic Variants (DGV). The genes overlapping CNVs that were different between twin pairs and called by at least three software programs were further characterized using Ingenuity Pathway Analysis (IPA; Ingenuity Systems, California) and GeneMania (Toronto, ON) to identify gene networks and canonical pathways. Finally, we compared the genes identified in this study to those genes listed in the Schizophrenia Gene Database (http://www.schizophreniaforum.org/res/sczgene/default.asp) to determine the genes that appear to be most likely to play a role in schizophrenia. Additional searches from PubMed covering the most recent results helped update any connection between genes of interest and disease pathology.
Confirmation of Unique CNVs
Differences between monozygotic twins were confirmed using TaqMan Quantitative PCR (qPCR) Copy Number Assays from Life Technologies. The control used for comparison of copy number in the TaqMan experiments was RNAse P and the calibrator used was the individual's unaffected co-twin.
Results
First, the number of unfiltered CNVs identified by each software program in each of the six twin pairs varied by program (Supplementary Table 1). Golden Helix's® SVS™ identified the highest number of CNVs in each individual, with a range of 168 to 209 variants. Affymetrix® Genotyping Console™ and Partek® Genomics Suite™ called similar numbers of CNVs in each individual, with ranges of 41 to 61 and 37 to 72 variants, respectively. PennCNV called the smallest number of CNVs in each individual, with a range of 21 to 53 variants. The smallest number of unfiltered variants called in any one individual was 21 (PennCNV) and the largest number was 209 (Golden Helix® SVS™). The majority of CNV calls across all four software programs were between 1 kb and 100 kb in size, with at least 56% of the total CNVs in each analysis falling into this range. Partek® Genomics Suite™ yielded the highest number of large calls (>10 Mb) and Golden Helix® SVS™ yielded the largest number of small CNV calls (1–100 kb).
TABLE 1 CNVs Identified by Affymetrix Genotyping Console, Partek Genomics Suite, and PennCNV (A/P/p), as Only Present in Affected Co-Twin

aChr = chromosome; bDGV = database of genomic variants.
Second, the percentage of Affymetrix® Genotyping Console™ CNV calls detected by three or more algorithms was 23.04%, the percentage of Partek® CNV calls detected by three or more algorithms was 30.30%, the percentage of PennCNV calls detected by three or more algorithms was 33.02% and the percentage of SVS™ CNV calls detected by three or more algorithms was only 0.36%. This summary of overlapping CNVs across four programs strongly suggests that our most reliable calls may represent overlapping CNVs involving Affymetrix® (A), Partek® (P), and PennCNV (p), termed as A/P/p. A combination that includes Golden Helix® along with any other two methods yielded rare CNVs only and was considered too restrictive. More importantly, the overlap generated by A/P/p calls was less restrictive across the twin pairs. Consequently, the CNVs identified by this combination were further assessed in the follow-up analysis involving shared and unshared CNVs between members of the six twin pairs studied (Supplementary Table 2).
TABLE 2 CNVs Identified by Affymetrix Genotyping Console, Partek Genomics Suite, and PennCNV (A/P/p), as Only Present in Unaffected Co-Twin

aChr = chromosome; bDGV = database of genomic variants.
Third, we found a total of 38 CNV events called by the three (A/P/p) software programs that were not shared with their co-twin across the six pairs. Specifically, 14 unique CNV events were observed in co-twins affected with schizophrenia (Table 1), while 22 were unique to the six unaffected co-twins of the six MZD pairs (Table 2). Some of the CNVs in both categories contained genes, while others were located in non-coding regions of the genome. In fact, there were a total of 12 unique genes overlapping the 15 CNVs that were found in affected members only. Similarly, there were a total of 28 unique genes overlapping the 23 CNVs that were found in unaffected twins only. The results confirm that monozygotic twins do differ for rare CNVs. They allow us to undertake pair-specific analysis in an effort to explain the discordance of the monozygotic twin pairs for schizophrenia, described below.
Twin Pair 1
The affected male patient in twin pair 1 was diagnosed with a psychotic disorder at age 19. He had 41, 53, and 36 raw CNV calls by each of the three (A, P, or p) programs, respectively. In comparison, his unaffected co-twin had 50, 72, and 41 raw CNV calls by the three methods. After CNV merging and discarding of CNVs that were not called by the combination of A/P/p, the affected and unaffected members of this twin pair yielded 14 and 12 CNVs, respectively. These CNVs fell into three categories; shared between the twin pair (10), unique to normal (2), and unique to the affected member of twin pair 1 (4). The two CNVs that were found to be unique to the unaffected twin were a loss at 2q22.1 and a gain at 7q35. These CNVs overlapped the genes THSD7B (Entrez Gene: 80731) and TPK1 (Entrez Gene: 27010), respectively (Table 2). Of the four CNVs that were found to be unique to the affected member, none are reported in the DGV. Three of these CNVs (6q14.1, 7q31.1, 14q21.2) cover no gene overlaps while one (17q21.31) covers the PYY (Entrez Gene: 5697) gene (Table 1). Interestingly, the 14q21 region has been previously implicated in bipolar disorder (Liu et al., Reference Liu, Juo, Dewan, Grunn, Tong, Brito and Baron2003). Also, PYY encodes a protein that has been previously identified to be a potential cerebrospinal fluid marker for mental illness (Widerlov et al., Reference Widerlov, Lindstrom, Wahlestedt and Ekman1988) and autism spectrum disorders (de Krom et al., Reference de Krom, Staal, Ophoff, Hendriks, Buitelaar, Franke and van Ree2009). This CNV loss for the PYY gene has not been previously reported in the DGV and may be a potential candidate for the discordance of this twin pair for schizophrenia.
Twin Pair 2
The affected female patient in twin pair 2 was diagnosed with schizoaffective disorder at age 27. She had 48, 66, and 41 raw CNV calls by each of the three (A, P, or p) programs, respectively. In comparison, her unaffected co-twin had 46, 65, and 42 raw CNV calls. After CNV merging and discarding of CNVs that were not called by the combination of A/P/p, 12 and 15 CNVs remained in the affected and unaffected twins, respectively. Of these, 11 were shared between the twin pair, 4 were unique to normal, and 1 was unique to the affected member of this twin pair. The four CNVs that were found to be unique to the unaffected member were a loss at 1q21.1 that overlapped two genes, a loss at 7q21.2 that overlapped no genes, a gain at 17q21.32 that overlapped six genes, and a loss at 19q13.13-19q13.2 that overlapped eight genes (Table 2). The CNV that was found to be unique to the affected member was a gain found in the region 11p15.1 that does not overlap with any known genes and has been previously reported in the DGV (Table 1). No CNVs identified in twin pair 2 have been reported for any neurodevelopmental disorder and the observations do not seem to be likely candidates to explain the discordance for schizophrenia seen in this twin pair.
Twin Pair 3
The affected female in twin pair 3 was diagnosed with paranoid schizophrenia at age 22. She had 47, 54, and 25 raw CNV calls by each of the three (A, P, or p) programs, respectively. In comparison, her twin sister had 44, 43, and 21 raw CNV calls. The merging and discarding of non-overlapping CNVs yielded 10 and 7 CNVs in the affected and unaffected twins, respectively. Of these, six of the CNVs were shared between twin pair 3, one was unique to normal, and four were unique to the affected member. The CNV that was found to be unique to the unaffected member was a loss at 3q26.1 that did not overlap any gene (Table 2). The four CNVs that were found to be unique to the affected twin overlapped four regions: 3p11.2-3p11.1, 7q11.21, 11p15.4, and 16p11.1 (Table 1). Of these, the loss at 3p11.2-3p11.1 is the only one that was not previously reported in the DGV. This CNV overlapped the EPHA3 (Entrez Gene: 2042) gene. This gene belongs to the ephrin receptor subfamily of protein-tyrosine kinases that have been implicated in mediating developmental events, particularly in the nervous system, and has been previously associated with neurodegenerative diseases (Martinez & Soriano, Reference Martinez and Soriano2005). Further, the 16p11 region has been implicated in mental disorders, including psychosis (Steinberg et al., Reference Steinberg, de Jong, Mattheisen, Costas, Demontis, Jamain and Stefansson2014). The other genes unique to the affected twin were LOC441242 (Entrez Gene: 441242), INTS4L2 (Entrez Gene: 644619), CCT6P1 (Entrez Gene: 643253), SNORA22 (Entrez Gene: 677807), OR52N2 (Entrez Gene: 390077), LOC283914 (Entrez Gene: 283914), LOC146481 (Entrez Gene: 146481), and LOC100130700 (Entrez Gene: 100130700). Of particular interest is OR52N2, an olfactory receptor. Olfactory receptors share a seven-transmembrane domain structure with many neurotransmitter and hormone receptors and are responsible for the recognition and G protein-mediated transduction of odorant signals (Malnic et al., Reference Malnic, Godfrey and Buck2004). Recent reports suggest robust olfactory deficits in schizophrenia patients (Moberg et al., Reference Moberg, Kamath, Marchetto, Calkins, Doty, Hahn and Turetsky2013). Yet another identified gene, SNORA22, encodes a small nucleolar ribonucleic acid (RNA), which may guide chemical modifications of other RNAs (Kiss, Reference Kiss2001). Interestingly, CCT6P1 is highly expressed in brain (Velculescu et al., Reference Velculescu, Zhang, Vogelstein and Kinzler1995). Also, the EPHA3 gene is the only CNV identified in this patient that has not been previously reported in DGV and has the potential to explain discordance for schizophrenia in this twin pair.
Twin Pair 4
The female patient in twin pair 4 was diagnosed with paranoid schizophrenia at age 18. She had 44, 49, and 25 raw CNV calls identified by each of the A, P, or p programs, respectively. In comparison, her unaffected co-twin had 52, 37, and 22 raw CNV calls. After CNV merging and discarding of CNVs that were not called by A/P/p, 10 and 7 CNVs remained in the affected and unaffected members of the twin pair, respectively; seven were shared between the twin pair and three were unique to the affected member. Interestingly, there was no CNV that was unique to the normal twin (Table 2). Of the three CNVs that were found to be unique to the patient, none are reported in the DGV. Two of the CNVs (3q25.1, 16q23.2) cover no genes while one (2q11.2) covers the KIAA1211L (Entrez Gene: 343990) gene (Table 1). Interestingly, the 3q25 region has been previously implicated in autism-spectrum disorders (Auranen et al., Reference Auranen, Vanhala, Varilo, Ayers, Kempas, Ylisaukko-Oja and Jarvela2002). Also, KIAA1211L is expressed in the brain and has been reported in bipolar disorder (Scott et al., Reference Scott, Muglia, Kong, Guan, Flickinger, Upmanyu and Boehnke2009). Also of interest, this CNV has not been previously identified in the DGV, making this CNV loss a potential candidate for the discordance for schizophrenia of this twin pair.
Twin Pair 5
The affected male in twin pair 5 was diagnosed with undifferentiated schizophrenia at age 20. He had 56, 54, and 45 raw CNV calls and his unaffected co-twin had 54, 58, and 41 such calls identified by the A, P, or p software programs, respectively. After CNV merging and discarding of CNVs that were not called by all three programs, 10 and 18 CNVs remained in the affected and unaffected twins, respectively. These CNVs fell into three categories: shared between the twin pair (8), unique to normal (10), and unique to the affected member of twin pair 5 (2). The 10 CNVs that were found to be unique to the unaffected member were found in the regions 1p36.33, 1p21.1, 1q25.2, 2p22.3, 3p14.1, 3q11.2, 4p15.1, 4q24, 8p11.23, and 9p11.2, and overlapped the PAPPA2 (Entrez Gene: 60676), EPHA6 (Entrez Gene: 285220), TACR3 (Entrez Gene: 6870), ADAM5 (Entrez Gene: 255926), LOC643648 (Entrez Gene: 643648), LOC283914, LOC146481, and LOC100130700 genes (Table 2). Of the two CNVs that were found to be unique to the patient (16p12.3 and 17q21.31), the first, a loss at 16p12.3, overlaps the GPR139 (Entrez Gene: 124274) gene and the second, a loss at 17q21.31, overlaps the PYY gene (Table 1). PYY, as presented in the pair-specific results shown for twin pair 1 above, is a potential candidate for the discordance for schizophrenia identified in this twin pair as well. GPR139 is a gene that is an important mediator of signal transduction. G-protein receptors are almost exclusively expressed in brain and are likely to play important roles in the central nervous system. GPR139 has been previously reported to be associated with attention deficit hyperactivity disorder (ADHD; OMIM: 143465; Ebejer et al., Reference Ebejer, Duffy, van der Werf, Wright, Montgomery, Gillespie and Medland2013). Both the CNV loss overlapping the GPR139 gene and the CNV loss overlapping the PYY gene have not been listed in the DGV. They may represent de novo events and candidates for the disease discordance of this twin pair.
Twin Pair 6
The male patient in twin pair 6 was diagnosed with paranoid schizophrenia at age 16. He had 61, 47, and 50 raw CNV calls as identified by the A, P, or p software programs, respectively. In comparison, his unaffected co-twin had 53, 48, and 53 raw CNV calls. After CNV merging and discarding of CNVs that were not called by all three (A/P/p) programs, 8 and 13 CNVs remained in the affected and unaffected members, respectively. These CNVs fell into three categories: shared between the twin pair (8), unique to normal (5), and unique to the affected member of twin pair 6 (0). The five CNVs that were found to be unique to the unaffected member were found at 2p22.3, 2q21.1, 3q26.1, 12p11.1, and 17q21.32. Only the CNV gains at 3q26.1 and 17q21.32 overlapped genes. The CNV at 3q26.1 overlapped the SPTSSB (Entrez Gene: 165679) gene and the CNV at 17q21.32 overlapped TTLL6 (Entrez Gene: 284076), CALCOCO2 (Entrez Gene: 10241), ATP5G1 (Entrez Gene: 516), and UBE2Z (Entrez Gene: 65264) (Table 2). No CNVs were found to be unique to the affected member of this twin pair. Consequently, no CNVs identified in twin pair 6 seem to represent candidates to explain their discordance for schizophrenia.
The results outlined above have identified rare and pair-specific CNV differences between monozygotic twins in each of the six twin pairs discordant for schizophrenia, studied. Some CNVs involve single or multiple genes and others represent non-coding genomic regions. Also, a number of these are not reported in DGV, specifically 10 of 14 events seen uniquely in affected twins and 9 of 22 events seen uniquely in unaffected members.
Discussion
It has become apparent that CNVs are common in human populations and play a significant role in the etiology of complex diseases, including schizophrenia (Ahn et al., Reference Ahn, Gotay, Andersen, Anvari, Gochman, Lee and Rapoport2013; St Clair, Reference St Clair2013). However, it is not easy to identify disease-specific CNVs and establish their mode of action in the causation of the disease. Of special concern is the use of arrays with different degrees of genome coverage and the large number of algorithms available to call CNVs. Although the Affymetrix Human Array 6.0 appears to meet most of the platform criteria, including coverage for CNV calling in humans, a gold standard algorithm for the analysis of data has not been established (Zhang et al., Reference Zhang, Qian, Akula, Alliey-Rodriguez, Tang and Liu2011). There is a likelihood of false positive results. Despite this, such experiments have generated and continue to generate valuable insights. Reports assessing the use of different software algorithms to analyze the same microarrays have identified a low concordance rate between software programs (Kim et al., Reference Kim, Kim and Chung2012; Pinto et al., Reference Pinto, Darvishi, Shi, Rajan, Rigler, Fitzgerald and Feuk2011). This is likely due to the substantial background noise, which contributes to a false discovery rate of variants (Grayson & Aune, Reference Grayson and Aune2011). To avoid this, often two programs are used to call for the CNVs and the resulting shared CNVs are considered to be reliable. Although logical, this approach is not totally satisfactory as it may ignore and miss out on some critical results. In this analysis we have focused on more reliable results and used four different software programs to call for CNVs. We found a low percentage of concordance between these calls. This is consistent with findings in the literature (Kim et al., Reference Kim, Kim and Chung2012; Pinto et al., Reference Pinto, Darvishi, Shi, Rajan, Rigler, Fitzgerald and Feuk2011) and highlights the necessity for more stringent guidelines for CNV calling from microarrays. A study conducted by Kim et al. (Reference Kim, Kim and Chung2012) suggested that at least three calling algorithms should be used to ensure the reliability of results.
As stated, we used four CNV calling programs (Golden Helix's® SVS™, Affymetrix® Genotyping Console™, Partek® Genomics Suite™, and PennCNV) and selected CNVs that were called by three methods (Affymetrix® Genotyping Console™, Partek® Genomics Suite™, and PennCNV) referred to as A/P/p. Also, we chose 10 CNVs to confirm the results by Real Time PCR. Our qPCR results established that one CNV, a CNV loss at 7q11.21 in twin pair 3 was significantly different between twins (Figure 2). Further, two CNVs (CNV loss at PYY in twin pair 5 and CNV gain at 5q11.2 in twin pair 6) showed the expected trend but were not statistically significant (Table 3). This suggests that the experimental confirmation by qPCR of the CNV calls is at least 10%.
TABLE 3 TaqMan Real Time PCR Results

*a significant difference between twin pair 3 in qPCR confirmation.

FIGURE 2 Copy number variation loss at 7q11.21 in twin 3A.
Yet another challenge with genetic studies in schizophrenia is the extensive heterogeneity that may include multiple genetic, epigenetic, and environmental factors (Singh & O'Reilly, Reference Singh and O'Reilly2009; van Dongen & Boomsma, Reference van Dongen and Boomsma2013). Two observations are of particular relevance to this discussion. First, a number of genomic regions and genes including CNVs, both inherited (relatively common) and de novo (extremely rare), have been implicated in this complex neurodevelopmental disease (Kirov, Reference Kirov2010; Maiti et al., Reference Maiti, Kumar, Castellani, O'Reilly and Singh2011; Van Den Bossche et al., Reference Van Den Bossche, Strazisar, Cammaerts, Liekens, Vandeweyer, Depreeuw and Del-Favero2013). Also, there is less than 100% concordance (48%) between monozygotic twins (McGuffin et al., Reference McGuffin, Asherson, Owen and Farmer1994). Consequently, the genome of the normal twin may provide a near perfect match to the genome of the affected member. Also, in some cases the discordance of monozygotic twins for schizophrenia could be attributed to differences in their CNVs potentially caused by DNM (Maiti et al., Reference Maiti, Kumar, Castellani, O'Reilly and Singh2011; Singh et al., Reference Singh, Castellani and O'Reilly2009). The published results suggest that DNM are not limited to the germ lines alone. Rather they are ongoing throughout life, including stages of differentiation, development, and aging (Lupski, Reference Lupski2010). The occurrence of DNMs has now been demonstrated using a variety of strategies including MZ twins (Bruder et al., Reference Bruder, Piotrowski, Gijsbers, Andersson, Erickson, de Ståhl and Dumanski2008; Singh et al., Reference Singh, Castellani and O'Reilly2009), trios (Vissers et al., Reference Vissers, de Ligt, Gilissen, Janssen, Steehouwer, de Vries and Veltman2010), and MZ twins compared with both parents (Maiti et al., Reference Maiti, Kumar, Castellani, O'Reilly and Singh2011). Quantitatively, Maiti et al. (Reference Maiti, Kumar, Castellani, O'Reilly and Singh2011) identified one and two DNMs in two pairs of MZ twins respectively, based on parental genotypes, while Vissers et al. (Reference Vissers, de Ligt, Gilissen, Janssen, Steehouwer, de Vries and Veltman2010) identified one to two DNMs per trio in eight trios with a mentally retarded proband using family-based exome sequencing. The results suggest that DNMs can account for phenotypic discordance between MZ pairs. Also, the degree of difference may vary from pair to pair. Their phenotypic impact will depend not only on the genomic region involved but also on the background genotype, and the timing of DNMs during ontogeny and the mechanism that is responsible for genomic discordance of MZ twins may generate mosaics, with or without significant phenotypic manifestation (Ruderfer et al., Reference Ruderfer, Chambert, Moran, Talkowski, Chen, Gigek and Ernst2013).
Our results on six MZD pairs show that each MZD pair differs in rare CNVs. Also, the discordance of some of the pairs could be attributed to CNVs identified in this analysis. For this, we have used the following criteria. First, the CNV should be present in the affected member(s) of the twin pair only. Second, the genomic region involved must encode for gene(s). Third, the gene must be expressed in the brain and/or the gene must have relevance to the neurodevelopment and physiological outcomes associated with schizophrenia. Furthermore, the CNV of interest must not have been identified previously in normal healthy individuals (DGV). The use of these criteria has allowed us to identify potential causes for schizophrenia in four of the six pairs studied. The four pairs that do meet our criteria have their own twin-pair-specific CNV patterns. Given extensive heterogeneity and the rare nature of de novo events, these patterns are expected to be variable. Not surprisingly, the observed differences are pair-specific with respect to the genomic region(s) and gene(s) involved. Although the results are patient and pair-specific, we did find some genomic regions and genes that are common across unrelated patients. For example, the region 16p11 was identified as uniquely disrupted in the affected member of twin pair 3 — this is particularly interesting as this region has been previously associated with psychosis (Steinberg et al., Reference Steinberg, de Jong, Mattheisen, Costas, Demontis, Jamain and Stefansson2014). Another example of a region identified in more than one sample was 2p22.3, the same CNV was uniquely identified in this region in the unaffected member of twin pair 5 and twin pair 6 — this may suggest a possible protective or mediating effect on the disease from a copy number variation loss in this region.
In conclusion, the MZD twin-based genomic (CNV) strategy to identify candidate genes that may be involved in schizophrenia is logical and practical. It has the potential to serve as an effective strategy in identification of genes and genetic mechanisms that may cause complex disorders. Specifically, individuals with schizophrenia have CNV gains and losses that are likely to contribute to the disease. Here, the inherited mutations may provide predisposition that may not be sufficient for disease manifestation. Occurrence of any/some additional de novo event (CNV or mutation) may add to this predisposition and manifest the disease. This two-hit model (Maynard et al., Reference Maynard, Sikich, Lieberman and LaMantia2001; Singh et al., Reference Singh, McDonald, Murphy and O'Reilly2004) of disease development may explain a number of observations on schizophrenia. First, in most cases of discordance in monozygotic twins, even the normal twin may have some or delayed manifestation of some or all symptoms. Second, in most cases of familial schizophrenia, the second hit may or may not be needed depending on the nature of familial predisposition. Also, in some cases environmental components may add to the predisposition or be enough by itself to affect neurodevelopment and result in the disease. What is needed is precise and reliable results that are not always forthcoming. This challenge is apparent from our results where only one of the 10 CNVs could be statistically confirmed, while two CNVs showed the expected trend but failed to reach the level of significance in qPCR. This follows a number of recent reports (Ehli et al., Reference Ehli, Abdellaoui, Hu, Hottenga, Kattenberg, van Beijsterveldt and Davies2012). Despite such limitations, the MZD strategy outlined appears realistic. Specifically, the use of strict criteria for the assessment of copy number variations in monozygotic twin genomes discordant for schizophrenia has identified a novel CNV (7q11.21) that is surrounded by low copy repeats with the potential to undergo mechanisms which generate CNVs de novo. This confirmed CNV was seen exclusively in the affected patient of twin pair 3 and deserves further investigation as a candidate region for schizophrenia and related disorders in this twin pair and beyond.
Supplementary Material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/thg.2014.6.
Acknowledgments
We wish to thank the individuals who participated in this study. We also wish to thank Beth Locke and Dr Mark Daley for their computational assistance. This work was supported by grants from the Canadian Institutes of Health Research (CIHR), Ontario Mental Health Foundation (OMHF), and Schizophrenia Society of Ontario. SS held Senior Research Fellowship of OMHF during the course of this research.







