Quantitative analysis of lateral root development with time-lapse imaging and deep neural network
Published online by Cambridge University Press: 13 February 2024
Abstract
During lateral root (LR) development, morphological alteration of the developing single LR primordium occurs continuously. Precise observation of this continuous alteration is important for understanding the mechanism involved in single LR development. Recently, we reported that very long-chain fatty acids are important signalling molecules that regulate LR development. In the study, we developed an efficient method to quantify the transition of single LR developmental stages using time-lapse imaging followed by a deep neural network (DNN) analysis. In this ‘insight’ paper, we discuss our DNN method and the importance of time-lapse imaging in studies on plant development. Integrating DNN analysis and imaging is a powerful technique for the quantification of the timing of the transition of organ morphology; it can become an important method to elucidate spatiotemporal molecular mechanisms in plant development.
Keywords
- Type
- Insights
- Information
- Creative Commons
- This is an Open Access article, distributed under the terms of the Creative Commons Attribution licence (https://creativecommons.org/licenses/by/4.0), which permits unrestricted re-use, distribution and reproduction, provided the original article is properly cited.
- Copyright
- © The Author(s), 2024. Published by Cambridge University Press in association with John Innes Centre
Footnotes
Associate Editor: Dr. Ari Pekka Mahönen
References

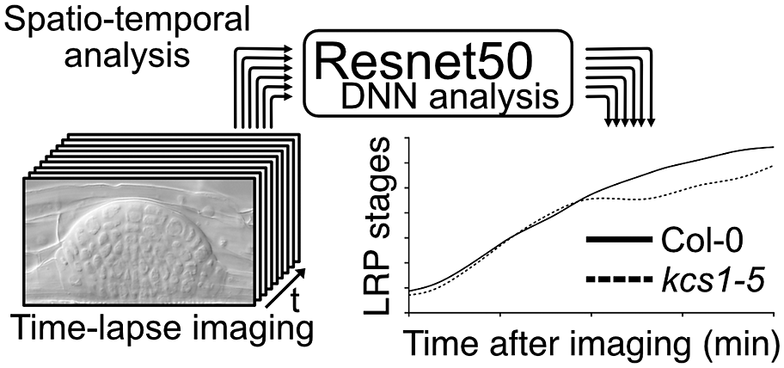
Figure 1. Comparison of conventional lateral root primordia (LRP) stage analysis methods (gravistimulation method) with a time-lapse imaging followed by a deep neural network (DNN) analysis method. (a) Schematic model of time-lapse imaging followed by the DNN analysis. LRP images obtained through time-lapse imaging serve as input for machine learning, facilitating the construction of a DNN model (in our case, utilising Resnet50). Subsequently, approximately 10 roots from experimental samples are subjected to time-lapse imaging to capture the developing LRP. By employing the established DNN model, automated identification of LRP stages is executed. The LRP stage transitions are then depicted graphically over time on the x-axis. Solid and dashed lines indicate LRP stage transition times for Col-0 and kcs1-5, respectively. The right bar graph presents statistical analysis results of DNN analysis at 2,760 min (46 h). The y-axis indicates the LRP stage determined using DNN. White box, Col-0; hatched box, kcs1-5. Significant difference from Col-0 was determined using the Welch’s t-test (**P = 0.004). The data were retrieved from Uemura et al. (2023). (b) Schematic model of the gravistimulation method. In each bio-replication experiment, at least 20 roots are subjected to gravity stimulation at a single time point. Following clearing of roots, the LRP stages are counted under a microscope. The right bar graph is the percentage of LRP at different stages of development after 46 h (2,760 min) of gravistimulation. Data are presented as mean ± SE of three biological replicates, with 20 seedlings in each replicate. White box, Col-0; hatched box, kcs1-5. Significant differences from Col-0 were determined using the generalised liner mixed model followed by Holm’s P-value adjustment in each stage (**P < 0.01, *P < 0.05). Similar to the bar graph in (a), the transition to the later stages of LRP development (stages VI and VII) was delayed in kcs1-5. The data were retrieved from Uemura et al. (2023). (c) Comparison of these two methods. One advantage of gravistimulation method is its high capacity to detect statistical differences. Conversely, a disadvantage is that as the number of time points, genotypes and treatments becomes enormous, data analysis becomes more complex. In the gravistimulation method, a minimal observation of at least 180 roots is required for a single genotype or treatment at a given time. The advantage of time-lapse DNN lies in conducting imaging with temporal information, requiring fewer individuals. With DNN, time-lapse imaging of 10 roots for a single genotype and treatment can be conducted, incorporating temporal information. However, a disadvantage is the need for pre-training the DNN with a substantial number of LRP images.
Author comment: Quantitative analysis of lateral root development with time-lapse imaging and deep neural network — R0/PR1
Comments
[20230728]
Editorial Team
Quantitative Plant Biology
Dear Editor:
I wish to submit an “insight” for publication in Quantitative Plant Biology titled “Quantitative analysis of lateral root development with time-lapse imaging and deep neural network.” The paper was coauthored by Yuta Uemura and Hironaka Tsukagoshi.
This insight paper aimed to discuss an effective technique to quantify the organ morphology using a combination of time-lapse imaging and deep neural network analysis. We believe that this paper will be of interest to the readership of your journal because time-lapse imaging can help elucidate the processes involved in plant development, and integrating it with DNN offers an effective technique to quantify the timing of the transition of organ morphology. Moreover, the technique we developed can help elucidate spatiotemporal molecular mechanisms in plant development.
All study participants provided informed consent, and the study design was approved by the appropriate ethics review board. We have read and understood your journal’s policies, and we believe that neither the manuscript nor the study violates any of these. There are no conflicts of interest to declare.
Thank you for your consideration. I look forward to hearing from you.
Sincerely,
Hironaka Tsukagoshi
Faculty of Agriculture
Meijo University
1-501 Shiogamaguchi, Tempaku-ku, Nagoya
Aichi, 468-8502 Japan
Review: Quantitative analysis of lateral root development with time-lapse imaging and deep neural network — R0/PR2
Conflict of interest statement
Comments
In the current insight, Uemura and Tsukagoshi make a case for quantitative analysis and scoring of lateral root development (in Arabidopsis). The authors make a case for the use of Deep Neural Networks (DNN) to score LRs in an automatic and non-supervised manner. They mostly refer to their recently published study (Uemura et al., 2023 TPJ) to make a case for this approach and propose that the use of DNN should be more explored for quantitative analysis of LR development (in this case) or any other developmental process. I think this insight would benefit from a figure that shows the different approaches one can use to study a given developmental process (confocal, cleared tissue, time lapse etc etc) and what the benefits/limitations are of each approach. This could give a clear insight as to what the power of the use of DNN is, but also for which traits it might not be so straightforward to implement a similar approach. I think this would make the insight a little bit more balanced and also more interesting for the general reader.
Review: Quantitative analysis of lateral root development with time-lapse imaging and deep neural network — R0/PR3
Conflict of interest statement
Comments
This manuscript is an “insight paper” – my understanding is that this is a short review of a key paper with additional insights and descriptions of how it connects to broader literature. This particular insight describes the work in Uemura et al, which used deep neural network (DNN) analysis to increase the speed of analysis of lateral root development in plants. The authors used their DNN pipeline to compare lateral root development in wildtype and mutants in the very-long chain fatty acid (VLCFA) pathway. The insight paper is interesting because it’s focus is very different from the original manuscript – the focus of the insight paper is on the DNN, but the original paper focuses much more on the biology. I think this is overall a strong approach, but there are some important ways that this paper could be improved.
1) It is not clear from reading this paper what the intention is. There is a brief mention in the abstract, but if you imagine someone downloading this paper off the internet without being familiar with “insight papers,” I think they might be very confused. To make the intention clearer, I think the introduction should include a description of the objective(s) of this manuscript. I also think that the main paper that is the focus of the review should be highlighted, instead of being cited like a normal paper. This will help it stand out to the reader.
2) Although I appreciate that the objective of this paper seems to be to focus on DNNs as a valuable research approach, there is a lot of information in the introduction about the biology of lateral root development. However, the biology discovered using a DNN is not really described (for instance, I had to read the source paper to learn that VLCFA is a regulator of LRP development through transcription factor-mediated regulation of gene expression and the transportation of VLCFAs is also involved in LR development through root cap cuticle formation). It’s a little bit of a disconnect. My recommendation is to match the introduction of the biology with a description of the results: so either shorten the biology descriptions in the introduction and focus instead on DNNs, or to have a longer summary of the biological results towards the middle/end of the manuscript.
3) There is a large gap between when DNN is introduced and lines 121 – 148, which describes why DNN was chosen for this type of analysis. I think the flow of the manuscript might be better if you raise this discussion earlier (maybe right after line 103, where you first introduce DNN).
4) Given that the focus of this manuscript is DNN analysis, I think it’s worth describing how the DNN performed compared to current gold standard methodology (how it was validated). This information is in the original paper, but a summary here would be useful. In addition, the source code could also be provided, for interested readers.
Minor comments:
There is need for editing for grammar and clarity throughout the document. Here are some important examples:
Line 59: recommend rewording to say “de novo tissue development” as opposed to “novel tissue development.”
Line 60: I’m not sure what the authors mean by “the lateral root temporally transitions between cellular and tissue states.” Perhaps adding citations would clarify this statement.
Line 64: I think that the authors are alluding to the fact that most labs study root development at a single time point, but especially for lateral roots, this is a dynamic process where time is an important component. This could be more clearly explained.
Line 97: The gravistimulated roots assay description should include a reference.
Line 101: This statement (“The combination of time-lapse imaging and …”) is a really critical part of the manuscript. This is where I think your central paper should be highlighted. The tense here is a bit strange – I think it might be clearer to say something along the lines of:
“We reasoned that the combination of time-lapse imaging and DNN analysis would make it possible to measure the developmental transitions of a single LRP. Therefore, we developed a machine learning-based method to quantify LRP development, as published in ‘A very long chain fatty acid responsive transcription factor, MYB93, regulates lateral root development in Arabidopsis’ (Uemura et al., The Plant Journal, 2023). Here, we describe the rational, development, and implications of the DNN approach for lateral root studies as well as explorations of many other biological questions.”
Recommendation: Quantitative analysis of lateral root development with time-lapse imaging and deep neural network — R0/PR4
Comments
Dear Dr. Tsukagoshi,
thank you for your submission to QPB. The reviewers found your study interesting, however, they found several issues that need to be improved. I hope that you find the comments useful.
Looking forward to read the revised version of your manuscript in the near future.
with best wishes,
Ari Pekka Mähönen
Decision: Quantitative analysis of lateral root development with time-lapse imaging and deep neural network — R0/PR5
Comments
No accompanying comment.
Author comment: Quantitative analysis of lateral root development with time-lapse imaging and deep neural network — R1/PR6
Comments
20231129
Editor-in-Chief
Dr. Olivier Hamant
Quantitative Plant Biology
Dear Editor:
We/I wish to re-submit the manuscript titled “Quantitative analysis of lateral root development with time-lapse imaging and deep neural network.” The manuscript ID is QPB-23 -0007.
We thank you and the reviewers for your thoughtful suggestions and insights. The manuscript has benefited from these insightful suggestions. I look forward to working with you and the reviewers to move this manuscript closer to publication in the Quantitative Plant Biology journal.
The manuscript has been rechecked and the necessary changes have been made in accordance with the reviewers’ suggestions. The responses to all comments have been prepared and attached herewith.
Thank you for your consideration. I look forward to hearing from you.
Sincerely,
Hironaka Tsukagoshi
Faculty of Agriculture, Meijo University, Nagoya, Aichi, Japan
E-mail: [email protected]
Review: Quantitative analysis of lateral root development with time-lapse imaging and deep neural network — R1/PR7
Conflict of interest statement
Comments
Overall, these review address my main concerns. I have one more suggestion:
Line 55: I’m still not exactly sure what the authors mean by “the LRP temporally transitions through cellular states and finally forms lateral root (Torres-Martínez et al., 2019).” I think this means that in comparison to the mature primary root, which maintains a distinct and stereotyped set of cell lineages in the meristem, the cells in the developing lateral root are dynamically undergoing changes in identity, at least until the root matures.
Review: Quantitative analysis of lateral root development with time-lapse imaging and deep neural network — R1/PR8
Conflict of interest statement
Comments
It serves its purpose as an insight. It would have been nice to have included a figure showing results obtained with the classical, gravistimulation method, and their DNN approach coupled to the live imaging. This could help the reader get insights into the results obtained when phenotyping the same genotypes with two different methods. In addition, it might be good to mention that the described approaches are mostly suited for Arabidopsis and perhaps not for species with larger roots or where gravistimulation might not work due to a more complex vascular system.
Recommendation: Quantitative analysis of lateral root development with time-lapse imaging and deep neural network — R1/PR9
Comments
Dear Prof. Tsukagoshi,
the two reviewers have now evaluated your manuscript, and overall the manuscript is getting close to acceptance. However, I agree with a reviewer that there is still a small textual issue (see details in reviewer’s comment) that needs to be handled before the acceptance.
best wishes,
Ari Pekka Mähönen
Decision: Quantitative analysis of lateral root development with time-lapse imaging and deep neural network — R1/PR10
Comments
No accompanying comment.
Author comment: Quantitative analysis of lateral root development with time-lapse imaging and deep neural network — R2/PR11
Comments
[20240116]
Olivier Hamant
Editor-in-Chief
Quantitative Plant Biology
Ari Pekka Mahönen
Associate Editor
Quantitative Plant Biology
Dear Editors:
We wish to re-submit the manuscript titled “Quantitative analysis of lateral root development with time-lapse imaging and a deep neural network analysis.” The manuscript ID is QPB-23-0007.R1.
We thank you and the reviewers for your thoughtful suggestions and insights. The manuscript has benefited from these insightful suggestions. We look forward to working with you and the reviewers to move this manuscript closer to publication in Quantitative Plant Biology.
The manuscript has been rechecked and the necessary changes have been made in accordance with the reviewers’ suggestions. The responses to all comments have been prepared and submitted along with the revised manuscript.
Thank you for your consideration. We look forward to hearing from you.
Sincerely,
Hironaka Tsukagoshi
Faculty of Agriculture, Meijo University, Nagoya, Aichi, Japan
Phone: +81-52-838-2372
E-mail: [email protected]
Recommendation: Quantitative analysis of lateral root development with time-lapse imaging and deep neural network — R2/PR12
Comments
No accompanying comment.
Decision: Quantitative analysis of lateral root development with time-lapse imaging and deep neural network — R2/PR13
Comments
No accompanying comment.
- 1
- Cited by



Elucidation of the molecular mechanisms through which root architecture is controlled is an important topic in plant development. Plant roots consist of primary root and lateral root (LR). For both root types, maintaining a balance between cellular proliferation and differentiation is key for adequate growth. The differentiation of LRs from the pericycle cells starting from lateral root primordia (LRP) formation can be considered a model case of novel tissue development. The primary root forms a series of cellular lineages from the root tip to the base; the cells in the developing LRP undergo dynamic changes in identity, at least until the LRP matures (Torres-Martínez et al., Reference Torres-Martínez, Rodríguez-Alonso, Shishkova and Dubrovsky2019). For such events, it is vital to control cellular function by regulating the function of phytohormones and their transcriptional in response to them (Banda et al., Reference Banda, Bellande, von Wangenheim, Goh, Guyomarc'h, Laplaze and Bennett2019; Fukaki & Tasaka, Reference Fukaki and Tasaka2009). Although extensive genetic and molecular analyses have revealed the molecular mechanisms of root architecture development, root development is a continuous process; therefore, spatiotemporal analysis must be performed.
Time-lapse imaging is a powerful tool used to reveal continuous root growth regulations because it enables the observation of spatiotemporal changes such as molecular signalling, organelle behaviour and organ development. To understand the regulation of target tissue development by genes, it is necessary to link quantitative phenotypic information with gene expression through time-series analysis. Therefore, a method for quantitatively analysing phenotypic changes and gene expression in the same time series with a high temporal resolution by observing gene expression and phenotypes in living plants is required.
In Arabidopsis thaliana, fluorescence time-lapse imaging of GCaMP, a fluorescent protein-based cytosolic [Ca2+] sensor, revealed momentary and dynamic Ca2+ signalling, which propagates to a leaf different from the one that accepts the signal at approximately 1 mm/s (Toyota et al., Reference Toyota, Spencer, Sawai-Toyota, Jiaqi, Zhang, Koo, Howe and Gilroy2018). Microscopic live tracking of organelle behaviour involving the process of root cap cell detachment, together with cell organelles, has revealed that the intracellular position of the nucleus changes during root cap detachment (Goh et al., Reference Goh, Sakamoto, Wang, Kozono, Ueno, Miyashima, Toyokura, Fukaki, Kang and Nakajima2022). In the case of organ development, confocal microscopy time-lapse imaging has provided direct evidence of the division patterns of stem cells and the proximal meristem in root tips (Campilho et al., Reference Campilho, Garcia, Toorn, Wijk, Campilho and Scheres2006). Furthermore, the cell division mechanism of LRP has been investigated by continuously observing the number of cells at each developmental stage of LRP, revealing differences in LRP development in individual plants (Campilho et al., Reference Campilho, Garcia, Toorn, Wijk, Campilho and Scheres2006; Lucas et al., Reference Lucas, Kenobi, von Wangenheim, Voβ, Swarup, De Smet, Van Damme, Lawrence, Péret, Moscardi, Barbeau, Godin, Salt, Guyomarc'h, Stelzer, Maizel, Laplaze and Bennett2013). It is critical to analyse and interpret continuous images captured via time-lapse imaging to elucidate seamless plant development. However, the number of images obtained from time-lapse imaging tends to be extremely high, and accurately discriminating and quantifying the growth of plants from such big data requires automation of image analysis. In this regard, analysis methods that apply machine learning are effective and indispensable. Digital image processing, including deep neural network (DNN) analysis, is an efficient and powerful technique for automatic image analysis. DNN analysis is superior to conventional digital image processing approaches in terms of computational programming simplicity. Conventional image recognition must quantify organ morphology through digital image processing, such as colour thresholding, feature extraction, statistics and algorithms. PlantCV provides a useful Python package for computational image processing that focuses on plant research (Fahlgren et al., Reference Fahlgren, Gehan and Baxter2015; Gehan et al., Reference Gehan, Fahlgren, Abbasi, Berry, Callen, Chavez, Doust, Feldman, Gilbert, Hodge, Hoyer, Lin, Liu, Lizárraga, Lorence, Miller, Platon, Tessman and Sax2017). To date, 181 digital image analysis tools, such as BRAT, a root length measurement tool, and GiA Roots, a root system architecture characterisation tool, have been developed; however, there are no DNN-based image processing tools with high contrast and background noise-reduction features (Galkovskyi et al., Reference Galkovskyi, Mileyko, Bucksch, Moore, Symonova, Price, Topp, Iyer-Pascuzzi, Zurek, Fang, Harer, Benfey and Weitz2012; Lobet, Reference Lobet2017; Lobet et al., Reference Lobet, Draye and Périlleux2013; Slovak et al., Reference Slovak, Göschl, Su, Shimotani, Shiina and Busch2014). None of the DNN-based image-processing methods comprises multiple algorithms; however, the DNN model is composed of neural network layers, and a combination of these layers determines the accuracy and application target. A DNN allows us to develop an image-recognition model by training an image dataset with high accuracy without complicated computer programming.
Phenotypic analysis of time-lapse images requires the quantification of the developmental stage, cell or tissue size, length, and population size. The automatic recognition of developmental stages from image data is a typical task in DNN applications. Various DNN classification models have been developed, such as Resnet50 and Xception, that can be used for stage classification without the need for complicated analytical pipelines (Chollet, Reference Chollet2017; He et al., Reference He, Zhang, Ren and Sun2016). Additionally, cell size, cell length and population size estimation can be used to perform DNN image segmentation and processing. DNN segmentation models, such as U-Net and X-Net, have been proposed (Fujii et al., Reference Fujii, Tanaka, Ikeuchi and Hotta2021; Ronneberger et al., Reference Ronneberger, Fischer and Brox2015). Each cell tissue size in a developed callus has been quantified through DNN image segmentation and image processing, and the DNN enables the quantification of different cell sizes (Ikeuchi et al., Reference Ikeuchi, Iwase, Ito, Tanaka, Favero, Kawamura, Sakamoto, Wakazaki, Tameshige, Fujii, Hashimoto, Suzuki, Hotta, Toyooka, Mitsuda and Sugimoto2022). PlantSeg has also been used for DNN segmentation and for cell length and volume quantification from Z-stack 3D images of roots and leaves captured using confocal laser microscopy (Graeff et al., Reference Graeff, Rana, Wendrich, Dorier, Eekhout, Aliaga Fandino, Guex, Bassel, De Rybel and Hardtke2021; Wolny et al., Reference Wolny, Cerrone, Vijayan, Tofanelli, Barro, Louveaux, Wenzl, Strauss, Wilson-Sánchez, Lymbouridou, Steigleder, Pape, Bailoni, Duran-Nebreda, Bassel, Lohmann, Tsiantis, Hamprecht, Schneitz and Kreshuk2020). These studies demonstrate that DNN models can be applied to other studies and obtain robust, accurate, and even efficient results for the quantification of plant development.
Recently, we reported that very long-chain fatty acids (VLCFAs) are involved in LR development through the regulation of the expression of the transcription factor MYB93 (Uemura et al., Reference Uemura, Kimura, Ohta, Suzuki, Mase, Kato, Sakaoka, Uefune, Komine, Hotta, Shimizu, Morikami and Tsukagoshi2023). In addition to MYB93, ATML1, which has a START domain (a lipid-binding region), reportedly binds to very long-chain ceramides and plays a key role in epidermal differentiation (Abe et al., Reference Abe, Katsumata, Komeda and Takahashi2003; Lu et al., Reference Lu, Porat, Nadeau and O'Neill1996; Sessions et al., Reference Sessions, Weigel and Yanofsky1999). pas2 mutants, which lack the function of VLCFA synthase, reportedly show abnormal cell proliferation in the shoot apical meristem (Nagata et al., Reference Nagata, Ishikawa, Kawai-Yamada, Takahashi and Abe2021; Nobusawa et al., Reference Nobusawa, Okushima, Nagata, Kojima, Sakakibara and Umeda2013). These results indicate that VLCFA synthesis and signalling are required for plant development. Additionally, the 3-ketoacyl-CoA synthase 1 (KCS1) gene encoding one of the key VLCFA synthesis enzyme is characteristically expressed in the LRP and LRP-peripheral cells, suggesting the involvement of VLCFAs in LRP development. Supporting these findings, several studies have shown that lower VLCFA levels affect LR development (Shang et al., Reference Shang, Xu, Zhang, Cao, Xin and Hu2016; Trinh et al., Reference Trinh, Lavenus, Goh, Boutté, Drogue, Vaissayre, Tellier, Lucas, Voß, Gantet, Faure, Dussert, Fukaki, Bennett, Laplaze and Guyomarc'h2019). We identified a transcription factor, MYB93, through RNA sequencing of a kcs1-5. The VLCFA levels decreased in kcs1-5 mutants, indicating that MYB93 is a novel transcription factor whose expression responds to VLCFA levels. MYB93 expression shows a specific response to fatty acid carbon chain length, with no response at C18 but a response to C20–C24 VLCFAs (Uemura et al., Reference Uemura, Kimura, Ohta, Suzuki, Mase, Kato, Sakaoka, Uefune, Komine, Hotta, Shimizu, Morikami and Tsukagoshi2023). Moreover, genetic analysis has revealed that MYB93 is involved in the late stages of LR development by regulating the expression of several cell wall remodelling genes, such as expansins (Uemura et al., Reference Uemura, Kimura, Ohta, Suzuki, Mase, Kato, Sakaoka, Uefune, Komine, Hotta, Shimizu, Morikami and Tsukagoshi2023).
Figure 1. Comparison of conventional lateral root primordia (LRP) stage analysis methods (gravistimulation method) with a time-lapse imaging followed by a deep neural network (DNN) analysis method. (a) Schematic model of time-lapse imaging followed by the DNN analysis. LRP images obtained through time-lapse imaging serve as input for machine learning, facilitating the construction of a DNN model (in our case, utilising Resnet50). Subsequently, approximately 10 roots from experimental samples are subjected to time-lapse imaging to capture the developing LRP. By employing the established DNN model, automated identification of LRP stages is executed. The LRP stage transitions are then depicted graphically over time on the x-axis. Solid and dashed lines indicate LRP stage transition times for Col-0 and kcs1-5, respectively. The right bar graph presents statistical analysis results of DNN analysis at 2,760 min (46 h). The y-axis indicates the LRP stage determined using DNN. White box, Col-0; hatched box, kcs1-5. Significant difference from Col-0 was determined using the Welch’s t-test (**P = 0.004). The data were retrieved from Uemura et al. (Reference Uemura, Kimura, Ohta, Suzuki, Mase, Kato, Sakaoka, Uefune, Komine, Hotta, Shimizu, Morikami and Tsukagoshi2023). (b) Schematic model of the gravistimulation method. In each bio-replication experiment, at least 20 roots are subjected to gravity stimulation at a single time point. Following clearing of roots, the LRP stages are counted under a microscope. The right bar graph is the percentage of LRP at different stages of development after 46 h (2,760 min) of gravistimulation. Data are presented as mean ± SE of three biological replicates, with 20 seedlings in each replicate. White box, Col-0; hatched box, kcs1-5. Significant differences from Col-0 were determined using the generalised liner mixed model followed by Holm’s P-value adjustment in each stage (**P < 0.01, *P < 0.05). Similar to the bar graph in (a), the transition to the later stages of LRP development (stages VI and VII) was delayed in kcs1-5. The data were retrieved from Uemura et al. (Reference Uemura, Kimura, Ohta, Suzuki, Mase, Kato, Sakaoka, Uefune, Komine, Hotta, Shimizu, Morikami and Tsukagoshi2023). (c) Comparison of these two methods. One advantage of gravistimulation method is its high capacity to detect statistical differences. Conversely, a disadvantage is that as the number of time points, genotypes and treatments becomes enormous, data analysis becomes more complex. In the gravistimulation method, a minimal observation of at least 180 roots is required for a single genotype or treatment at a given time. The advantage of time-lapse DNN lies in conducting imaging with temporal information, requiring fewer individuals. With DNN, time-lapse imaging of 10 roots for a single genotype and treatment can be conducted, incorporating temporal information. However, a disadvantage is the need for pre-training the DNN with a substantial number of LRP images.
In an analysis of LRP development under the regulation of VLCFAs and MYB93, we developed a machine learning-based method for quantifying LRP development. LRP development has conventionally been assessed by inducing LRP development and analysing the distribution of developmental stages by observing the LRP at several time points. In general, the gravistimulation assay has been used widely to count LRP stages (Lee et al., Reference Lee, Cho and Kim2015; Péret et al., Reference Péret, Li, Zhao, Band, Voß, Postaire, Luu, Da Ines, Casimiro, Lucas, Wells, Lazzerini, Nacry, King, Jensen, Schäffner, Maurel and Bennett2012). In this assay, gravistimulated roots are fixed at several time points, and the LRP developmental stages are counted under a microscope. This analysis provides information on LRP development-stage distribution in bulk samples. Therefore, a single LRP development cannot be traced, and there is a lack of information on the transition time among different LRP stages. We deduced that the combination of time-lapse imaging and DNN analysis would enable to measure the developmental transitions of a single LRP. Furthermore, conducting multi-time point experiments with different genotypes or chemical treatments increases the sample size significantly even within a single experiment, placing a substantial burden on researchers. Therefore, we developed a machine learning-based method for quantifying LRP development, as published in ‘A very long chain fatty acid responsive transcription factor, MYB93, regulates LR development in Arabidopsis’ (Uemura et al., Reference Uemura, Kimura, Ohta, Suzuki, Mase, Kato, Sakaoka, Uefune, Komine, Hotta, Shimizu, Morikami and Tsukagoshi2023). Here, we describe the rationale, development and implications of the DNN approach for LR studies, as well as explore numerous other biological questions. We performed LRP time-lapse imaging using VLCFA-related mutants, such as kcs1-5, and image analysis using the DNN. For LRP time-lapse imaging, we developed a system that could capture time-lapse images under a microscope while the plants are growing on the medium and monitored tissue development over several days. However, high time-resolution time-series analysis makes it difficult to quantify phenotypes because it requires processing a large number of images. LRP development is classified into stages I–VII based on morphological features. Therefore, it requires the manual annotation of thousands of LRP time-lapse images of different stages, times and genotypes. We solved this problem by using DNNs to automatically quantify phenotypes, using Resnet50 as the DNN model that was developed by training approximately 8,000 images to create this model (Figure 1). This method captures the transition time between LRP stages. The assessment of LRP development using time-lapse image analysis showed that developmental delay occurs in the later stages of LRP development in kcs1-5 and MYB93 overexpressors (Figure 1). In this study, we also conducted LR induction experiments via gravistimulation and compared the results using the DNN analysis. kcs1-5 and MYB93 overexpressors exhibited reduction in the number of LRP in the later stages, consistent with the DNN analysis results (Figure 1). In the gravistimulation assay, the number of LRP in at least 180 roots was measured under one condition, with biological replicates of three sets of 20 roots at three different time points for a single genotype or chemical treatment. Therefore, when considering mutants, overexpressors, and each chemical treatment, the total number exceeds 1,000 roots. Combined LRP time-lapse imaging and DNN phenotypic analysis quantitatively indicated the phenotypic changes over time from only 20 roots. Moreover, using our system, we could classify LRP stages in not only mutants but also plants subjected to any chemical treatment. Compared with the gravistimulation assay, our method offers major advantages in terms of reducing labour, improving stage discrimination accuracy and increasing research speed. However, to construct a DNN model, it is imperative to initially acquire a substantial number of images. In our case, after acquiring over 8,000 images of various LRP stages, we classified the images manually and subsequently employed machine learning techniques. Once the training phase is validated, the analysis of newly captured time-lapse images becomes feasible with considerably fewer images. Moreover, it is important to analyse and interpret continuous images captured via time-lapse imaging to elucidate seamless plant development. We have shown that it is effective to quantify LRP development using a DNN with time-lapse imaging. Even in time-lapse imaging, this technique is useful for studying other tissues and cells. This is because it enables the observation of spatiotemporal changes such as molecular signalling, organelle behaviour and organ development along with the imaging of certain reporters. However, our analysis was conducted only in Arabidopsis. For other plant species with larger root tissues than Arabidopsis, optimisation of imaging and other preparations might be essential.
Time-lapse imaging is a tool for capturing a substantial number of images over time. The development and growth processes of plants undergo continuous changes, making it necessary to analyse morphological changes accurately by considering multiple time points. However, a gap persists owing to a lack of technique for the automatic quantification and statistical processing of phenotypic information. In plants, deep learning has been leveraged for tasks such as classification and segmentation. This facilitates the automated identification of disease species through the analysis of images depicting plant diseases and enumeration of leaf numbers based on shoot images (Singh et al., Reference Singh, Ganapathysubramanian, Sarkar and Singh2018; Ubbens & Stavness, Reference Ubbens and Stavness2017; Yuan et al., Reference Yuan, Xu, Zhai and Xu2023). The application of deep learning has streamlined the quantification of phenotypic information from images, which was previously challenging. Through the quantitative analysis of extensive image datasets, including time-lapse LRP images, insights can be gleaned from the wealth of temporal information.
Acknowledgements
We would like to thank Editage for English language editing.
Funding statement
This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) KAKENHI (Grant No. JP23H04207).
Competing interest
The authors declare no competing interests.
Author contributions
This manuscript was conceived and written by Y.U. and H.T.
Data availability statement
No data or code were developed for this manuscript.