INTRODUCTION
Norovirus (NoV) has historically been designated ‘winter vomiting diseases’, later as ‘Norwalk virus’ and ‘Norwalk-like virus’, and finally as ‘norovirus’ [Reference Adler and Zickl1, Reference Lopman, Zambon and Brown2]. NoV can be classified into five genogroups (GI–GV) by phylogenic analysis of the capsid protein. Human NoV infections are caused by GI, GII, and, to a lesser extent, GIV [Reference Wang3]. Genogroups GI and GII of human NoV are further classified into 14 and 17 subgenotypes, respectively [Reference Cheon4]. Symptoms of NoV include acute-onset vomiting, watery diarrhoea, and nausea. Healthy adults usually recover without complications [Reference Mattner5]. NoV can be transmitted efficiently due to the low infectious dose and the high viral load in faeces and vomitus [Reference Repp and Keene6].
Historically, NoV outbreaks were under-reported; however, with development of reverse transcription–polymerase chain reaction (RT–PCR)-based diagnostics, NoV has been recognized as a leading cause of gastroenteritis worldwide [Reference Kirkwood7]. Over-reporting of disease is also possible due to a lack of proof that NoV detected by PCR is viable, and able to cause illness or be transmitted. In the USA, NoV is the most common cause of foodborne illness outbreaks. The primary mode of transmission is person-to-person, followed by foodborne, environmental contamination, and waterborne transmission [Reference Hall8]. As many as 19–21 million NoV cases, including 56 000–71 000 hospitalizations, and 570–800 deaths, are reported annually in the USA [Reference Hall9].
Outbreaks of gastroenteritis (defined as ⩾2 cases of a similar illness) in South Korea are reported to the Korea Centers for Disease Control and Prevention (KCDC), and NoV was designated in South Korea as a national notifiable infectious disease in 2006. During 2007–2009, NoV was the most common pathogen identified in foodborne disease outbreaks, occurring most frequently from November to May [Reference Gwack10].
In February 2012, a NoV outbreak in elementary school A was reported. In succession, another NoV outbreak occurred in elementary school B, about 20 km from school A. We conducted epidemiological investigations to characterize the extent of the outbreaks, identify the mode of transmission, and determine the relationship between the outbreaks.
METHODS
The outbreaks
On 4 February 2012, a school nurse from school A reported to a local health authority that more than 20 students had diarrhoea or vomiting. On 15 February, a local clinic reported to another local health authority an outbreak of gastroenteritis of more than ten students from school B. These outbreaks were reported to the KCDC; each local health authority first separately initiated an epidemiological investigation. After the common features of these outbreaks became apparent, the KCDC assumed responsibility for the epidemiological investigations.
Case definition and study design
Cases were defined as illness in students with diarrhoea (⩾3 loose stools in any 24-h period) or vomiting (at least one episode). Standardized self-administered questionnaires were used to elicit information on demographic characteristics, clinical history, and potential exposures (food, water, milk, snacks). In school A, all students were enrolled and a retrospective cohort study was undertaken. In school B, suspected students experiencing symptoms of loose stool or vomiting were detected in cooperation with teachers. A case-control study was conducted using healthy controls that were two randomly selected students in the same class. We calculated the relative risk (RR) or odds ratio (OR) with 95% confidence intervals (CI) to assess associations between illness and potential exposures. These public health investigations undertaken to control outbreaks did not require approval by an institutional review board.
Site inspection
The cafeterias and kitchens in schools A and B were separately inspected for environmental exposures to the pathogens on 2 February and 15 February, respectively. Inspection of suspected company X was undertaken on 17 February to review the green seaweed production process.
Laboratory testing
Students who met the case definition as well as food handlers were encouraged to provide stool samples. Samples of preserved food items served on 2 February in school A and 13 February in school B and environmental samples from kitchen knives, chopping boards, and dishcloths were collected. Drinking and tap water was also tested. From company X, samples of green seaweed (1 kg), seawater (40 litres) used for washing raw green seaweed, and seawater (40 litres) collected near company X were obtained to test for NoV.
Enteric pathogens were examined as described previously [Reference DuPont11]. NoV was identified using conventional nested RT–PCR [Reference Cheon4]. All PCR products were sequenced using an ABI Prism 3100 automatic sequencer and Big Dye Terminator Cycle Sequencing Mix (Invitrogen, USA). The 185 bp of NoV capsid genes were aligned with ClustalW and compared to those of reference strains obtained from GenBank. Phylogenetic analysis was conducted using the neighbour-joining method with 1000 bootstrap replicates in MEGA, version 4.0 [Reference Cheon4].
RESULTS
Epidemiological investigation
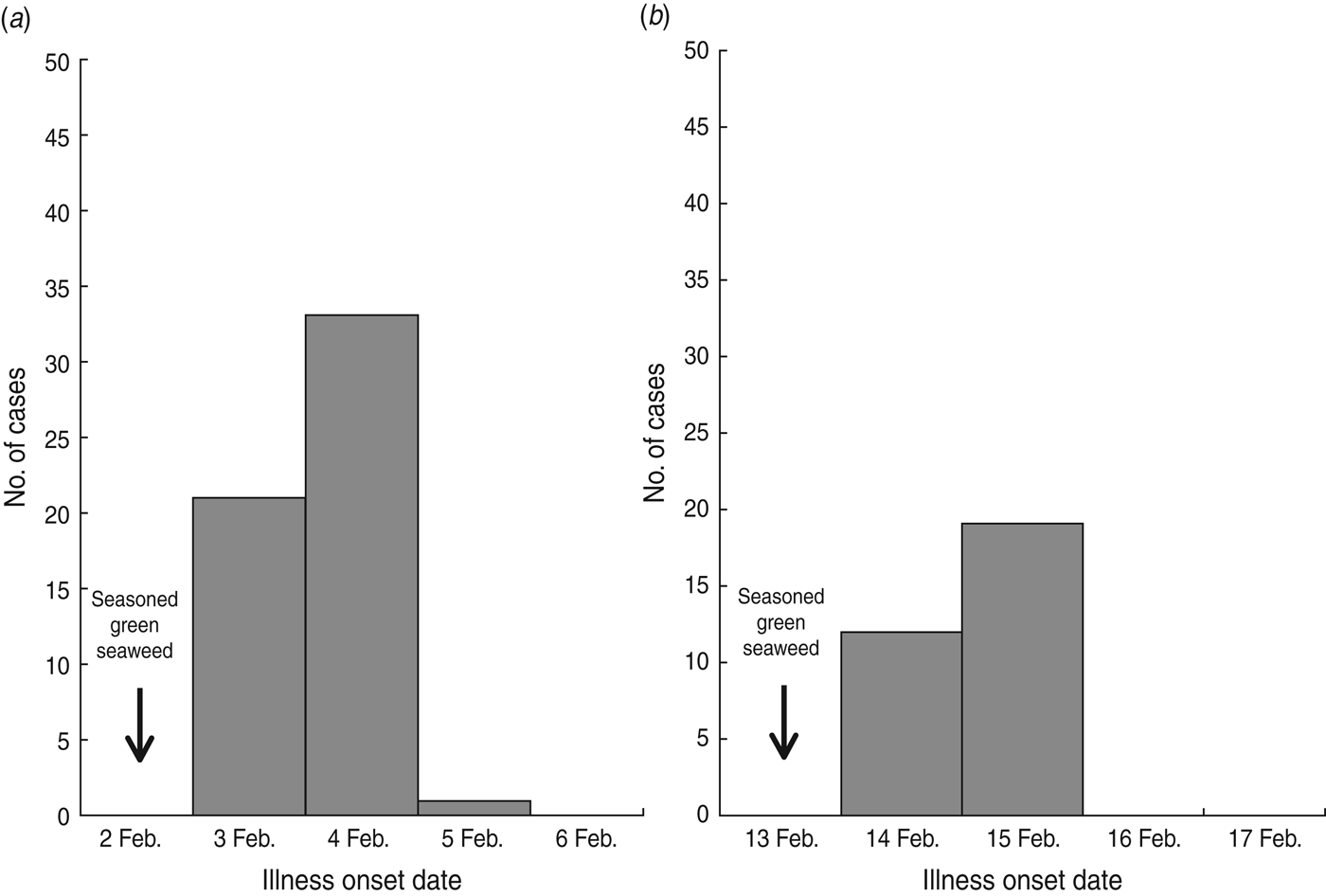
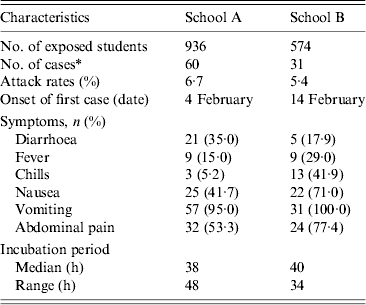
Of 936 students in school A, 893 (response rate 95·4%) participated in the retrospective cohort study. Sixty pupils (attack rate 6·7%) met the case definition. The primary symptoms were vomiting (95·0%), abdominal pain (53·3%), and nausea (41·7%) (Table 1). The outbreak onset occurred in the early morning of 3 February, with the number of cases peaking the following morning (Fig. 1 a).

Fig. 1. Epidemic curves of norovirus outbreaks in (a) school A and (b) school B.
Table 1. General characteristics of norovirus outbreaks in the two schools

* Cases were defined as illness with diarrhoea (⩾3 loose stools in any 24-h period) or vomiting (at least one episode) in students.
Of 574 pupils in school B, 31 (attack rate 5·4%) of 39 suspected student cases met the case definition. The most common symptom was vomiting (100·0%), followed by abdominal pain (77·4%) and nausea (71·0%) (Table 1). The first case displayed symptoms early in the morning of 14 February, and the peak in the number of cases occurred the next morning (Fig. 1 b). The median incubation period was 38 h (range 14–62 h) from lunch on 2 February and 40 h (range 14–48 h) from lunch on 13 February in schools A and B, respectively (Table 1). The attack rates in different grades showed no difference in schools A and B.
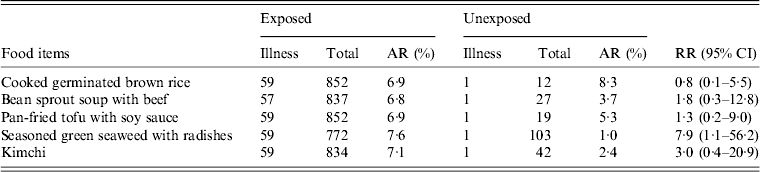
On 1 February, school A opened after the winter vacation and there were no major events or visits to other places from 1 January to 5 February. Lunch had been served on weekdays since 2 February. Among lunch food served on 2 February, uncooked green seaweed, and seasoned vinegar served with sliced radishes, was significantly associated with illness (RR 7·9, 95% CI 1·1–56·2) (Table 2). There was no common exposure of snacks or drinking water (brought from home) in cases.
Table 2. Association of illness with lunch food served on 2 February in school A

AR, Attack rate; RR, relative risk; CI, Confidence interval.
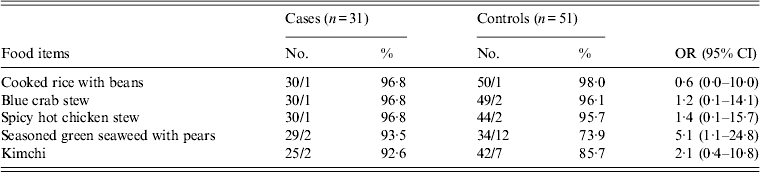
In school B, lunch was served on weekdays and there were no major events or visits to other places from 1 January to 15 February. Among lunch food served on 13 February (Monday), cases were 5·1 times more likely to have eaten uncooked green seaweed served with vinegar seasoning and sliced pears (95% CI 1·1–24·8) (Table 3). Other food items, cafeteria drinking water, and snacks were not significantly associated with illness. Tap water was used for cooking and none of food handlers in schools A or B reported symptoms. During the investigations, we observed proper food handling practices and hygiene practices in schools A and B.
Table 3. Association of illness with lunch food served on 13 February in school B

OR, Odds ratio; CI, Confidence interval.
The food items served in schools A and B were seasoned green seaweed and kimchi. Kimchi, a traditional fermented food in South Korea, consists of vegetables and a seasoning mixture that includes spices and minor additives. The green seaweed used by both schools came from company X, located on the coastal area of Busan. The kimchi, however, was sourced from different food companies. Company X obtained raw green seaweed from more than two farms. The seaweed was washed with seawater at company X to remove foreign matter. Company X then provided green seaweed to school A (2 February) and school B (13 February).
Laboratory testing
In school A, stool samples were available from 19 cases and seven food handlers. Fifteen stool samples (78·9%) from cases and four stool samples (57·1%) from food handlers were positive for NoV. In school B, stool samples were obtained from 30 cases and eight food handlers. Five stool samples (16·7%) from cases were positive for NoV. Stool samples from food handlers were negative for NoV.
Samples of drinking and tap water from schools A and B were negative for routine tests for general bacteria, total coliforms, and Escherichia coli. Samples of preserved food items and environmental samples from kitchen knives, chopping boards, and dishcloths from schools A and B were negative for bacterial pathogens, but were not tested for viral pathogens including NoV. Samples of green seaweed, seawater used for washing, and seawater collected near company X were positive for NoV.
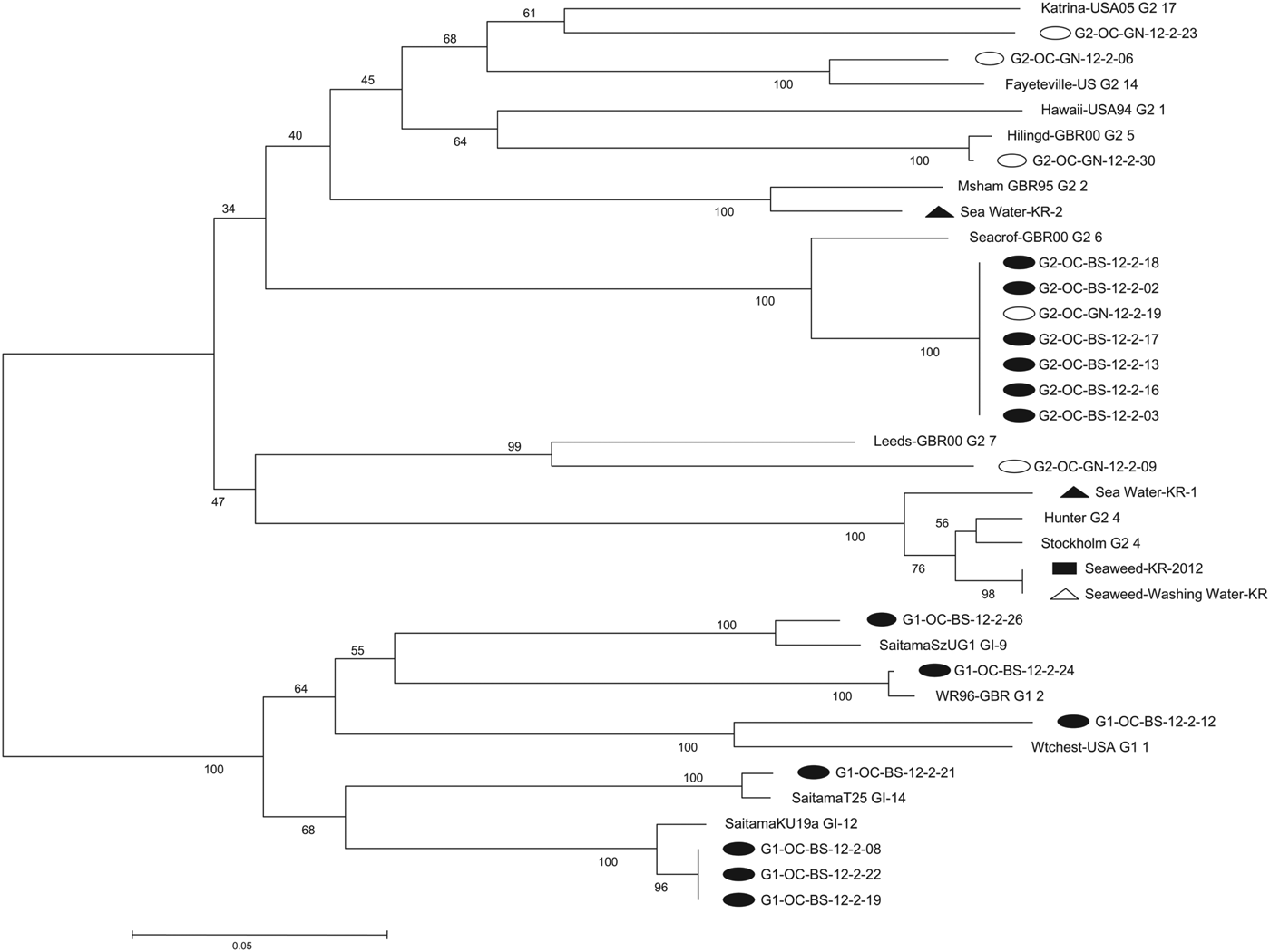
Phylogenetic analysis (Fig. 2)
Phylogenetic analysis was used to evaluate the relatedness of NoV strains detected in the two outbreaks and company X. The genotypic distribution of the 18 NoV strains isolated from the two outbreaks was GII.6, 38·9% (n = 7); GI.12, 16·7% (n = 3); GII.5, 5·6% (n = 1); GII.7, 5·6% (n = 1); GII.14, 5·6% (n = 1); GII.17, 5·6% (n = 1); GI.1, 5·6% (n = 1); GI.2, 5·6% (n = 1); GI.9, 5·6% (n = 1); GI.14, 5·6% (n = 1). School A yielded 13 NoV strains belonging to six genotypes including GI.1, GI.2, GI.9, GI.12, GI.14, and GII.6. School B yielded five NoV strains belonging to five genotypes including GII.5, GII.6, GII.7, GII.14, and GII.17. NoV GII.6 samples from the two outbreaks were indistinguishable.

Fig. 2. Phylogenetic analysis of outbreaks based on partial sequence of the norovirus capsid region (open reading frame 2). Nucleotide sequences were analysed by the neighbour-joining method. The numbers at the branches indicate bootstrap values for 1000 replicates. Closed circles, 13 strains detected in school A; open circles, five strains detected in school B; closed rectangle, one strain detected in green seaweed from company X; closed triangles, two strains detected in seawater near company X; open triangle, one strain detected in seawater used for washing raw green seaweed from company X.
Company X yielded four NoV strains of two genotypes: GII.4 (n = 3; green seaweed, seawater used for washing, and seawater near company X) and GII.2 (n = 1; seawater near company X). NoV GII.4 from green seaweed and seawater used for washing was clustered and showed 98% sequence homology.
DISCUSSION
The most common symptom associated with the outbreaks in these elementary schools was vomiting. Vomiting is more frequently reported than diarrhoea in children diagnosed with NoV [Reference Sala12]. The median incubation periods were 38 h and 40 h in these two outbreaks, consistent with the reported average incubation period of 24–48 h [Reference Patel13]. Seasoned green seaweed was significantly associated with illness in both NoV outbreaks, and company X provided green seaweed to both schools. Phylogenetic analysis revealed that NoV GII.6 was indistinguishable between the two outbreaks. Samples obtained from company X of green seaweed and seawater used for washing were positive for NoV GII.4. They showed 98% sequence homology in phylogenetic analysis. No additional NoV outbreaks were identified in prospective surveillance of adjacent schools, clinics, and hospitals.
NoV can be transmitted from person to person [Reference Hall8]; however, the epidemic curves had one peak in each outbreak and we found no evidence of person-to-person transmission among pupils, teachers, and food handlers between school A and school B, which are located in different provinces. Additionally, multiple NoV genotypes were identified in the two outbreaks, a finding consistent with sewage contamination or foodborne contamination earlier in the food-handling chain. Besides person-to-person transmission, the possibility of waterborne transmission was low because there was no common drinking water or cooking water between schools A and B.
Green seaweed belongs to the genus Enteromorpha, which occurs commonly along the North European and Mediterranean shores, and is distributed broadly throughout the coasts of South Korea, China, and Japan. Green seaweed resides in the upper region of the intertidal zone or in eutrophicated coastal water. It has been used for millennia in Asia as a pharmaceutical product and healthcare food, and have previously been utilized in the USA, France, and Chile [Reference Cho14–Reference Li16].
Among food items served in schools A and B, only seasoned green seaweed and kimchi were uncooked; NoV could survive in both these food items [Reference Wang and Tian17]. Green seaweed was supplied from company X, while kimchi was supplied from different companies. The green seaweed in this study was eaten unheated along with vinegar seasoning. While the vinegar could eliminate some microbes, including E. coli, NoV remains stable in these acidic conditions [Reference Chang and Fang18–Reference Lopman20].
A study of freshwater and seawater in Europe using nested RT–PCR reported NoV-positive rates of 6·3% and 16·4% in freshwater and seawater, respectively [Reference Wyn-Jones21]. In South Korea, the NoV-positive rate using nested RT–PCR was higher (84·4%) in freshwater from four rivers [Reference Lee and Kim22], implying a marked potential for contaminated seafood. Shellfish play an important role in the transmission of NoV due to the accumulation by filtration of large volumes of contaminated seawater [Reference Suffredini23], and a correlation between NoV concentration in wastewater and in oysters has been reported [Reference Flannery24].
Company X and at least one green seaweed farm are located on the southern coast of Korea near the Nakdong River, where wastewater flows into the sea. The NoV GII.4 genotypes detected in samples of green seaweed and seawater used for washing were not identical. NoV GII.4 was not isolated from samples obtained from these two outbreaks. We were unable to determine if the NoV contamination occurred at the farms or during washing at company X. However, the green seaweed sample from company X was positive for NoV, and could be the source of infection in these two NoV outbreaks.
To our knowledge, this is the first report of norovirus outbreaks associated with consumption of green seaweed; however, the findings in this study have several limitations. First, the preserved seasoned green seaweed in the two schools could not be tested for NoV owing to small volumes. Second, we could not conduct a retrospective cohort study of the outbreak in school B due to limited time and resources. Third, asymptomatic infections might dilute the association between illness and the consumption of food items. We reviewed the possibility of other contaminated food items as a cause of these outbreaks; however, only consumption of uncooked green seaweed could explain these NoV outbreaks. Fourth, although the food handlers in school A reported no symptoms, four food handlers were positive for NoV. However, these food handlers had eaten the same seasoned green seaweed provided to the NoV-positive cases.
After the outbreaks, company X was forbidden from selling green seaweed. We reported a summary of these outbreaks in the KCDC newsletter to inform the public of the risks of consuming unheated seasoned green seaweed [25]. A seafood monitoring system that includes green seaweed, oysters, freshwater, and seawater might be considered to decrease NoV infections.
ACKNOWLEDGEMENTS
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
DECLARATION OF INTEREST
None.