Introduction
Itchgrass [Rottboellia cochinchinensis (Lour.) Clayton] is recognized as one of the most noxious and troublesome annual weeds in tropical and subtropical regions (Bolfrey-Arku et al. Reference Bolfrey-Arku, Chauhan and Johnson2011; Leon et al. Reference Leon, Izquierdo and González-Andújar2015). In the Americas, it is frequently found from the Gulf of Mexico to the Caribbean regions of Central and South America (Leon et al. Reference Leon, Izquierdo and González-Andújar2015), where it causes serious problems, including yield reduction, in several annual crops such as beans (Phaseolus vulgaris L.), cassava (Manihot esculenta Crantz.), cotton (Gossypium hirsutum L.), maize (Zea mays L.), peanut (Arachis hypogaea L.), pineapple [Ananas comosus (L.) Merr.], sugarcane (Saccharum officinarum L.), and rice (Oryza sativa L.), and in perennial crops such as banana (Musa sp.), citrus (Citrus spp.), mango (Mangifera indica L.), and oil palm (Elaeis guineensis Jacq.) at early growth stages (Bolfrey-Arku et al. Reference Bolfrey-Arku, Chauhan and Johnson2011).
Over the last decade, acetyl-coenzyme A carboxylase (ACCase)-inhibiting herbicides, which selectively inhibit the homomeric plastidic form of ACCase (EC.6.4.1.2) (Kaundun Reference Kaundun2014), have been extensively used for managing R. cochinchinensis POST in a variety of crops (Avila et al. Reference Avila, Bolaños and Valverde2007; Heap 2014, Reference Heap2017). However, populations of R. cochinchinensis in Bolivia (Avila et al. Reference Avila, Bolaños and Valverde2007), Costa Rica (Castillo-Matamoros et al. Reference Castillo-Matamoros, Brenes-Angulo, Herrera-Murillo and Gómez-Alpízar2016), and the United States (Heap Reference Heap2017) have evolved resistance to ACCase inhibitors, including fluazifop-P-butyl, haloxyfop-R-methyl, fenoxaprop, and cyhalofop-butyl from the APP group, and clethodim and sethoxydim from the cyclohexanedione (CHDs/DIMs) chemistry. Resistance to these herbicides has made the management of this important weed more difficult and expensive. Therefore, a reliable, rapid, and simple diagnosis of resistance is necessary to maintain herbicide efficacy by preventing further spread of herbicide-resistant plants and improving the implementation of integrated resistance management strategies (Beffa et al. Reference Beffa, Figge, Lorentz, Hess, Laber, Ruiz-Santaella and Strek2012; Burgos et al. Reference Burgos, Tranel, Streibig, Davis, Shaner, Norsworthy and Ritz2013).
Resistance to ACCase herbicides, as with other herbicide modes of action, is widely divided into target-site (TSR) and non–target site (NTSR) resistance mechanisms. TSR is due to target-gene mutations, while NTSR is due to enhanced rates of herbicide metabolism (metabolic resistance) (Kaundun Reference Kaundun2014).
Different approaches have been used to determine resistance to ACCase herbicides, including conventional whole-plant pot assay and molecular methods (Burgos Reference Burgos2015; Burgos et al. Reference Burgos, Tranel, Streibig, Davis, Shaner, Norsworthy and Ritz2013; Délye et al. Reference Délye, Jasieniuk and Le Corre2013; Singh et al. Reference Singh, Sharma, Raghav, Chhokar and Sharma2015). The former method is the most widely used and reliable test to assess herbicide resistance (Burgos Reference Burgos2015; Burgos et al. Reference Burgos, Tranel, Streibig, Davis, Shaner, Norsworthy and Ritz2013). However, it presents several disadvantages, such as being labor-intensive, time- and space-consuming, and it takes at least 30 d to provide results and can show variation depending on application conditions and genotype by environment interactions. In addition, it cannot definitively identify the type of resistance (i.e., whether it is due to mutations or a physiological mechanism) (Beffa et al. Reference Beffa, Figge, Lorentz, Hess, Laber, Ruiz-Santaella and Strek2012). Molecular methods, on the other hand, could overcome most of these drawbacks, allowing for quick and precise detection of novel and known mutations associated with TSR, enabling the development of high-throughput herbicide-resistance diagnostics (Beffa et al. Reference Beffa, Figge, Lorentz, Hess, Laber, Ruiz-Santaella and Strek2012; Burgos et al. Reference Burgos, Tranel, Streibig, Davis, Shaner, Norsworthy and Ritz2013; Délye et al. Reference Délye, Jasieniuk and Le Corre2013; Singh et al. Reference Singh, Sharma, Raghav, Chhokar and Sharma2015). Non–target site metabolic resistance is more difficult to determine.
TSR to ACCase-inhibiting herbicides is based on point mutations (single-nucleotide polymorphisms [SNPs]) occurring at seven codon positions in the carboxyl-transferase (CT) domain of plastid-localized ACCase that prevent or reduce herbicide binding (Jang et al. Reference Jang, Marjanovic and Gornicki2013; Kaundun Reference Kaundun2014; Liu et al. Reference Liu, Harrison, Chalupska, Gornicki, O’Donnell, Adkins, Haselkorn and Williams2007). To date, 14 nonsynonymous mutations, (Ile-1781 to Leu/Val/Arg/Thr, Trp-1999 to Cys/Leu/Ser, Trp-2027 to Cys, Ile-2041 to Asn/Val, Asn-2078 to Gly, Cys-2088 to Arg, and Gly-2096 to Ala/Ser) within the CT domain have been reported as conferring resistance (Beckie and Tardif Reference Beckie and Tardif2012; Kaundun Reference Kaundun2014). Recently, resistance to fluazifop-P-butyl in R. cochinchinensis has been demonstrated to be due to a Trp-to-Cys substitution at position 2027 of the plastidic ACCase. This mutation arises from a change of a guanine (G) residue to a cytosine (C) at the third position in the cognate tryptophan codon (Castillo-Matamoros et al. Reference Castillo-Matamoros, Brenes-Angulo, Herrera-Murillo and Gómez-Alpízar2016).
Currently, the most common molecular method used for detection of SNPs associated with resistance to ACCase-inhibiting herbicides is the amplification of the CT domain by polymerase chain reaction (PCR), followed by sequencing for confirmation (Burgos et al. Reference Burgos, Tranel, Streibig, Davis, Shaner, Norsworthy and Ritz2013; Kaundun Reference Kaundun2014). However, both the labor and costs involved in PCR-DNA sequencing are limiting factors for routine application of this method for herbicide-resistance detection (Burgos et al. Reference Burgos, Tranel, Streibig, Davis, Shaner, Norsworthy and Ritz2013). Several alternative methods, including cleaved amplified polymorphic sequence (Kaundun et al. Reference Kaundun, Hutchings, Dale and McIndoe2012; Scarabel et al. Reference Scarabel, Panozzo, Savoia and Sattin2014) and derived cleaved amplified polymorphic sequence (Délye et al. Reference Délye, Pernin and Michel2011; Kaundun and Windass Reference Kaundun and Windass2006), both forms of PCR–restriction fragment-length polymorphism and PCR of specific alleles (PASA) (Délye et al. Reference Délye, Matéjicek and Gasquez2002; Raghav et al. Reference Raghav, Singh, Chhokar, Sharma and Kumar2016), PCR–single-strand conformation polymorphism (Singh et al. Reference Singh, Sharma, Raghav, Chhokar and Sharma2015), and real-time PCR (RT-PCR) (Kaundun et al. Reference Kaundun, Cleere, Stanger, Burbidge and Windass2006), have been developed for allele-specific detection enabling the discrimination of resistant and susceptible individuals. Further methods like loop-mediated isothermal amplification (Pan et al. Reference Pan, Li, Zhang and Dong2015), pyrosequencing (Hess et al. Reference Hess, Beffa, Kaiser, Laber, Menne and Strek2012; Petersen et al. Reference Petersen, Dresbach-Runkel and Wagner2010), and SNaPshot assay (Alarcón-Reverte et al. Reference Alarcón-Reverte, Hanley, Kaundun, Karp and Moss2013) have also been developed; however, most of these methods require post-PCR handling (separation of PCR products in a gel or other matrix to detect sequence variation), making these less suitable for high-throughput analysis, in addition to being costly and able to only a few specific mutations (Burgos Reference Burgos2015; Burgos et al. Reference Burgos, Tranel, Streibig, Davis, Shaner, Norsworthy and Ritz2013).
High-resolution melting analysis (HRMA) is a method that detects SNPs in target regions without the need for sequencing (Reed et al. Reference Reed, Kent and Wittwer2007). It is simple, rapid, sensitive, effective, cost-effective, and closed-tube (no post-PCR handling needed) (Reed et al. Reference Reed, Kent and Wittwer2007; Simko Reference Simko2016; Tong and Giffard Reference Tong and Giffard2012); thus it might be suitable as a quick diagnostic method for herbicide resistance. Furthermore, HRMA is a nondestructive method, which means that subsequent analysis of the sample by other methods like DNA sequencing or gel electrophoresis is possible (Liu et al. Reference Liu, Wu, Yang, Xu, Chen, Huang and Fu2014). The method involves PCR amplification of the mutation-containing region in the presence of a saturating double-stranded DNA (dsDNA)-binding dye followed by a melting (denaturation/dissociation into single-stranded DNA [ssDNA]) step. The dye shows high levels of fluorescence when bound (dsDNA), but has minimal fluorescence in the unbound (ssDNA) state. Therefore, the addition of the dye to the PCR reaction results in the generation of fluorescence proportional to the amount of the dsDNA amplicon present in the reaction tube. After PCR is completed, the amplicon (typically 50- to 200-bp long) is gradually denaturated by increasing the temperature in small increments (ca. 0.01 to 0.2 C). The temperature increase causes a progressive decrease in the fluorescence signal as the two strands of dsDNA melt apart, which is monitored in real time and plotted (fluorescence intensity vs. temperature) to produce a characteristic melting profile (melting analysis) for the amplicon. The melting profile is dependent on GC content, length, sequence and complementary of DNA strands (Reed et al. Reference Reed, Kent and Wittwer2007; Simko Reference Simko2016; Tong and Giffard Reference Tong and Giffard2012) and the melting temperature at which 50% of the DNA is in the double-stranded state can be approximated by taking the derivative of the melting curve (Liu et al. Reference Liu, Wu, Yang, Xu, Chen, Huang and Fu2014).
HRMA has been successfully applied for screening of SNPs in genes associated with antibiotic (mainly in human pathogens) (Nagai et al. Reference Nagai, Iwade, Hayakawa, Nakano, Sakai, Mitarai, Katayama, Nosaka and Yamaguchi2013; Tong and Giffard Reference Tong and Giffard2012), fungicide (Chatzidimopoulos et al. Reference Chatzidimopoulos, Ganopoulos, Madesis, Vellios, Tsaftaris and Pappas2015; Curvers et al. Reference Curvers, Pycke, Kyndt, Vanrompay, Haesaert and Gheysen2015; Samaras et al. Reference Samaras, Madesis and Karaoglanidis2016), and insecticide (Pasay et al. Reference Pasay, Arlian, Morgan, Vyszenski-Moher, Rose, Holt, Walton and McCarthy2008) resistance. To the best of our knowledge, no prior study has evaluated HRMA for diagnosis and screening mutations involved in ACCase-inhibitor resistance. Therefore, the objective of this study was to develop an HRMA assay that effectively and consistently detected the Trp-2027-Cys mutation associated with ACCase-inhibitor resistance in R. cochinchinensis.
Materials and Methods
Primer Design
A pair of primers was designed using Primer3Plus program (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) based on a 292-bp CT domain of the ACCase gene sequence alignment from fluazifop-P-butyl–susceptible and fluazifop-P-butyl–resistant R. cochinchinensis plants (GenBank accession numbers: KM592092 and KM592093; Castillo-Matamoros et al. Reference Castillo-Matamoros, Brenes-Angulo, Herrera-Murillo and Gómez-Alpízar2016): RottF: 5′-GTTGACCCAGCCTGAAGAAT-3′ and RottR: 5′-GGATTGCCTCTGTTCATCCT-3′. Primers amplify an 89-bp fragment containing the target polymorphic site TGG (Trp)/TGC (Cys), codon 2027 (according to blackgrass [Alopecurus myosuroides Huds.] numbering, GenBank accession no. AJ310767); responsible for the mutation Trp-2027-Cys, associated with resistance to ACCase-inhibiting herbicides (Figure 1). Primers were synthesized by Macrogen (Seoul, South Korea).

Figure 1 Sequence alignment showing the single-nucleotide change (G/C) within the chloroplastic acetyl-coenzyme A carboxylase gene carboxyl-transferase domain fragments from Rottboellia cochinchinensis susceptible (G) and resistant (C) biotypes. The sequences of the HRMA primers are colored in red. Position numbers (Alopecurus myosuroides full ACCase sequence, GenBank AJ310767, numbering) are given above the nucleotide sequences. Conserved nucleotides are indicated by dots.
DNA Extraction
Total genomic DNA was extracted from fresh leaf tissue of 3-wk-old herbicide-resistant and herbicide-susceptible seedlings, as described in Castillo-Matamoros et al. (Reference Castillo-Matamoros, Brenes-Angulo, Herrera-Murillo and Gómez-Alpízar2016). The DNA samples were then diluted to 1:5 (10 ng ul−1 working concentration) and stored at −20 C until further use.
RT-PCR HRMA
The RT-PCR reaction mixture (10 μl total volume) contained 5 μl of 2X HRM PCR master mix, including the ds-DNA–binding fluorescent EvaGreen dye (Type-it HRM PCR Kit, Qiagen, Hilden, Germany), forward (RottF) and reverse (RottR) primers (0.7 μl each of 10 μM working stocks), 2 μl genomic DNA, and 2.3 μl RNAse-free water. The reactions were set up in a Rotor-Gene Q RT-PCR system (Qiagen) under the following conditions: 95 C for 5 min, followed by 40 cycles of 10 s at 95 C, and then 30 s at 55 to 60 C. HRMA was performed immediately following the completion of PCR amplification. The temperature was raised from 65 to 85 C at 0.1 C increments with a 2-s hold time for each acquisition step. Rotor Gene Q software (Qiagen) was used to set up the sample arrangement, to define PCR conditions, for monitoring the amplification in real time, and for viewing and analyzing the melting curves.
To study the ability of the HRMA assay to detect heterozygous variants present in an R. cochinchinensis population, artificial heterozygotes (heteroduplex) were created by mixing PCR products (post-PCR mixing) of wild-type and mutant homozygotes using a mixing ratio of 1:1 (Cheng et al. Reference Cheng, Yim, Low, Tay and Yap, Lai2011) to simulate heterozygous genotype. All three genotypes were subjected to HRMA as described earlier.
Sequencing of PCR Products
To verify the HRMA results, DNA samples were PCR amplified using the same HRMA primers. PCR amplification was performed in a final volume of 25 μl including 2 μl of crude DNA extract, 2.5 μl 10X DreamTaq PCR reaction buffer (Thermo Fisher Scientific, Waltham, MA), 1 μl of each primer (10 μM), 2 μl of nucleotide mix (2 mM), 1.7 μl of MgCl2 (25 mM), 1.0 μl of bovine serum albumin (20 mg ml−1), and 0.25 μl of DreamTaq polymerase (5 U/μl) (Thermo Fisher Scientific), with ddH2O added to a final volume of 25 μl. The cycling conditions were DNA denaturation for 30 s at 95 C, 37 cycles of 10 s at 95 C, 15 s at 60 C (first set of primers) or 61 C (second set of primers), and 45 s at 72 C; and finally, a 10-min extension time at 72 C. The amplification was checked in a 1.6% agarose gel containing GelRed and visualized under UV light. PCR products were sequenced according to the Sanger method by Macrogen in two directions using the PCR primers. Sequences were aligned and compared using BioEdit Sequence Alignment Editor Software (Hall Reference Hall1999).
Quantification of HRMA Accuracy
DNA extraction and analysis was done from 10 susceptible and 10 resistant R. cochinchinensis individuals based on the fluazifop-P-butyl–resistance characterization previously done by Castillo-Matamoros et al. (Reference Castillo-Matamoros, Brenes-Angulo, Herrera-Murillo and Gómez-Alpízar2016). After DNA extraction and sequencing as described earlier, DNA samples were blindly analyzed to identify fluazifop-P-butyl–resistant and fluazifop-P-butyl–susceptible plants using HRMA. Sequencing and HRMA results were compared to quantify HRMA accuracy.
Results and Discussion
The primers used successfully amplified all samples tested with conventional PCR (Figure 2) and produced a single band of 89 bp. No nonspecific bands were observed. Robust, specific PCR is one of the main determinants for a successful application of HRMA for SNP detection (Reed et al. Reference Reed, Kent and Wittwer2007). PCR products were kept short (89 bp), because the size of the amplicon and GC content have a significant impact on the accurate discrimination and identification of alleles. PCR amplicons of relatively small length increased the melting temperature (Tm) shift between two sequences that differed at only one nucleotide position, thereby making them easier to differentiate (Curvers et al. Reference Curvers, Pycke, Kyndt, Vanrompay, Haesaert and Gheysen2015; Gundry et al. Reference Gundry, Dobrowolski, Martin, Robbins, Nay, Boyd, Coyne, Wall, Wittwer and Teng2008). Although there are multiple primers available in the literature to amplify domains with mutations known to confer herbicide resistance (Délye et al. 2002, Reference Délye, Pernin and Michel2011), it is likely that those primers will not be appropriate to conduct HRMA, because they amplify relatively long DNA sequences (Liew et al. Reference Liew, Pryor, Palais, Meadows, Erali, Lyon and Wittwer2004; Reed et al. Reference Reed, Kent and Wittwer2007). Therefore, to apply this method to other species and genes, specific primers for sequences close to the mutation of interest, such as those used in the present study, should be designed to increase the ability of the HRMA to distinguish between resistant and susceptible individuals.
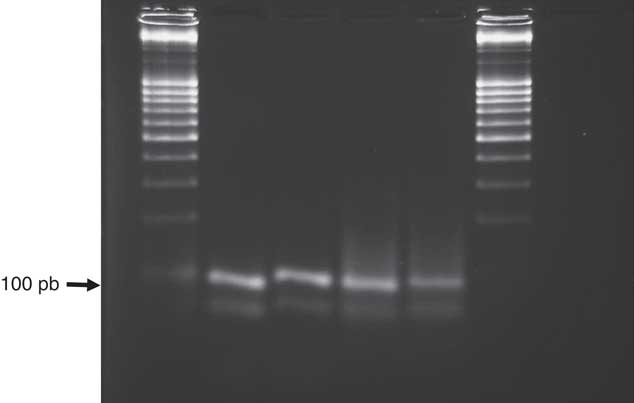
Figure 2 Agarose gel (1,8%) showing polymerase chain reaction products (89 bp) of the chloroplastic acetyl-coenzyme A carboxylase gene carboxyl-transferase domain targeted by HRMA primers RottF and RottR.
With HRMA, a combination of differences in Tm, normalized curve shape, and difference plots were used to discriminate sequence variation. The amplicons with a saturating fluorescent dye were melted to generate melting curves (Figures 3a and 4a), normalized plots (Figures 3b, 4b, and 5a), and difference plots (Figures 3c, 4c, and 5b). The latter shows the relative melting curves with reference to the susceptible (wild-type, TGG) biotype (Figures 3c, 4c, and 5b, horizontal blue line). The fluazifop-P-butyl–resistant R. cochinchinensis biotype (mutant with the C sequence) was clearly differentiated from the susceptible biotype (wild type with the G sequence). Both homozygous biotypes (susceptible and resistant) produced melt curves that were similar in shape, but the PCR amplicon from resistant biotype melted at a higher temperature than the susceptible biotype (80.78 vs. 80.37 C). This Tm difference (0.41 C) is considered significant, because it lies in the range of 0.2 to 0.5 C expected for a G-to-C transversion (class III SNP) (Curvers et al. Reference Curvers, Pycke, Kyndt, Vanrompay, Haesaert and Gheysen2015).
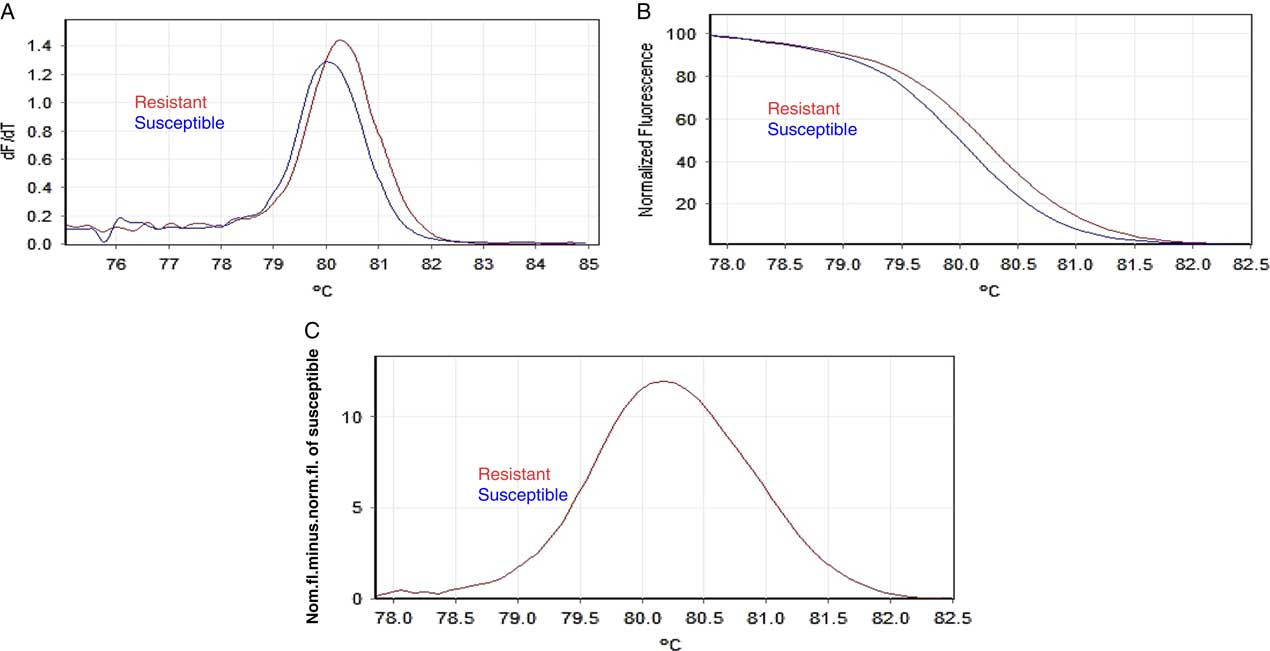
Figure 3 High-resolution melting analysis (HRMA) for detection of mutation Trp-2027-Cys in the Rottboellia cochinchinensis carboxyl-transferase domain of the acetyl-coenzyme A carboxylase gene conferring resistance to fluazifop-P-butyl. Representative profiles of the melting curves (derivative melt curves) (A), normalized melt curves (B) and differential curves using susceptible (wild type) as reference genotype (C) for resistant (TGC, in red) and susceptible plants (wild type, TGG in blue at x-axis level).
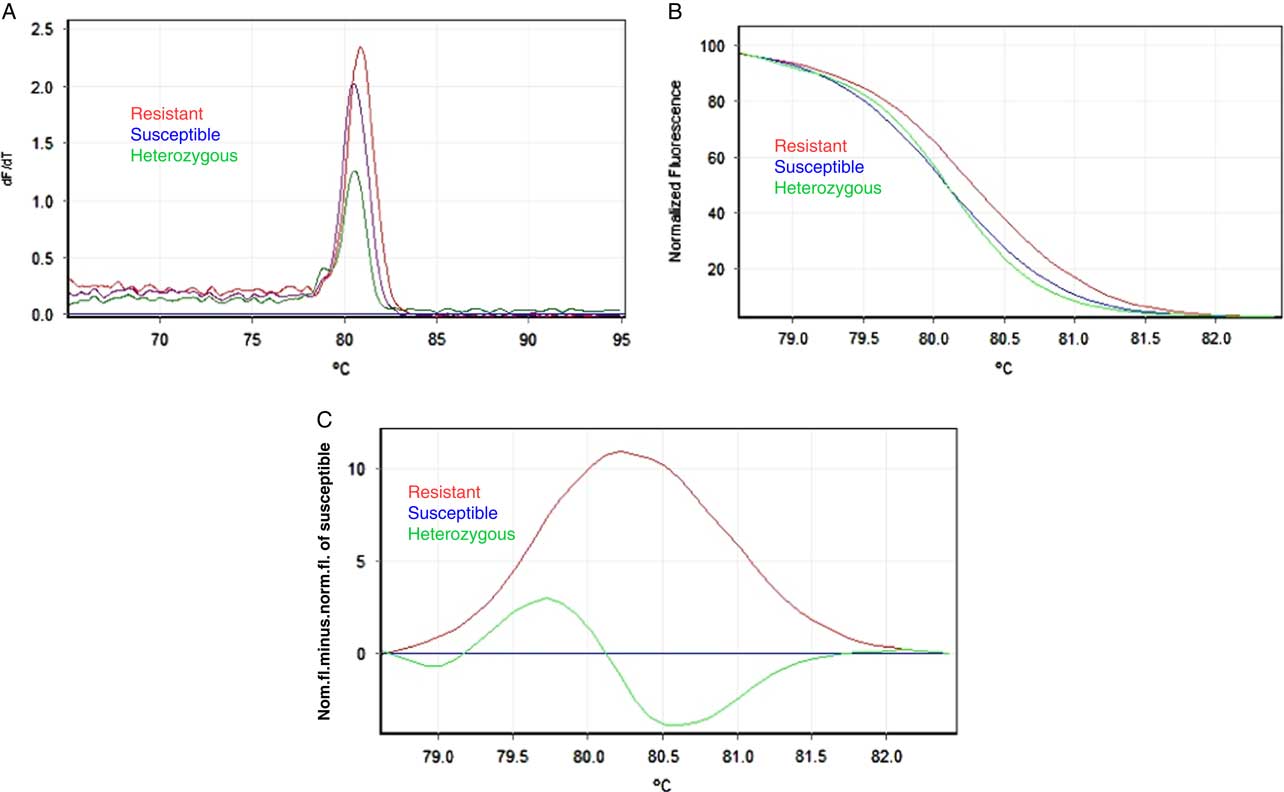
Figure 4 High-resolution melting analysis (HRMA) for detection of mutation Trp-2027-Cys in Rottboellia cochinchinensis carboxyl-transferase domain of the acetyl-coenzyme A carboxylase gene conferring resistance to fluazifop-P-butyl. Three genotypes are included: wild type (homozygous TGG, susceptible), mutant homozygous (TGC, resistant), and artificial mutant heterozygous (TGG/TGC, possibly resistant). (A) Representative profiles of the melting curves (derivative melt curves), (B) normalized plot, and (C) difference plot using susceptible (wild type) as the reference genotype.
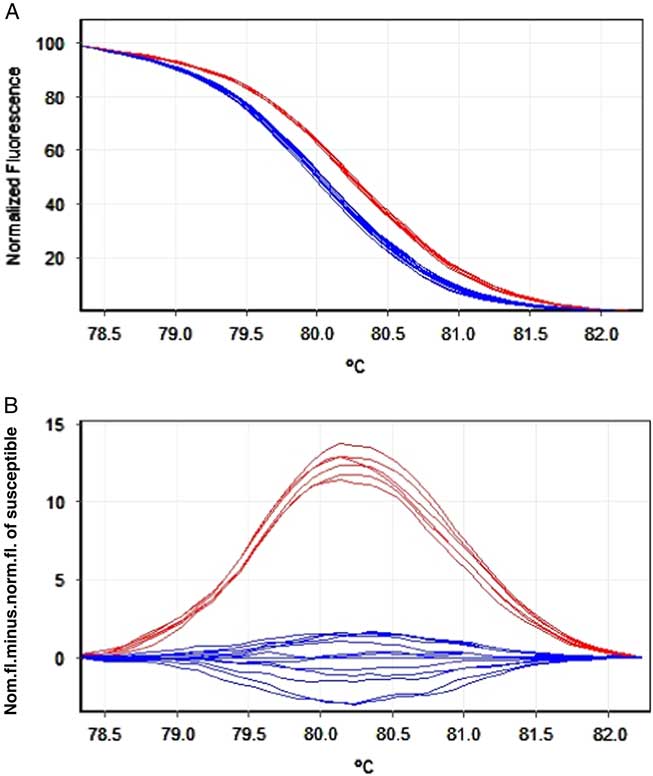
Figure 5 High-resolution melting analysis (HRMA) for detection of the Trp-2027-Cys mutation in Rottboellia cochinchinensis carboxyl-transferase domain of the acetyl-coenzyme A carboxylase gene conferring resistance to fluazifop-P-butyl. (B) Normalized plot and (C) difference plot using susceptible (wild type) as the reference genotype.
On the other hand, the artificial heterozygote was distinguished by a change in melting curve shape and slope (Figure 4b). The differential plot enhances visualization of the curve shape in an easier way, and the three genotypes were easily distinguished (Figure 4c). To date, point mutations occurring at seven codon positions in the CT domain of chloroplastic ACCase lead to ACCase-inhibitor resistance. Resistant plants may carry one or several of these mutations in the homozygous or heterozygous states (Kaundun Reference Kaundun2014); however, to date, only homozygous 2027-Cys fluazifop-P-methyl–resistant R. cochinchinensis plants have been detected (Castillo-Matamoros et al. Reference Castillo-Matamoros, Brenes-Angulo, Herrera-Murillo and Gómez-Alpízar2016). Furthermore, this does not preclude the presence of heterozygous resistant plants with the 2027 allele in the population, as has been shown for this mutation in other resistant grass weeds (Délye et al. Reference Délye, Matéjicek and Michel2008).
Our findings were in agreement with the detection sensitivity of HRMA regarding heterozygous variants (Cheng et al. Reference Cheng, Yim, Low, Tay and Yap, Lai2011). Therefore, the HRMA we developed should discriminate between homozygous and heterozygous R. cochinchinensis plants for the mutant 2027 allele. Although a validation with real heterozygous samples is needed, previous research has demonstrated that identification of heterozygotes is easy and consistent with HRMA, but depending on the sequence of the amplicon, the presence of heterozygotes could create an apparent homozygote signal (Liew et al. Reference Liew, Pryor, Palais, Meadows, Erali, Lyon and Wittwer2004). However, because most target-site mutations for herbicide resistance are dominant, this potential error does not prevent the detection of the resistant allele. Discrimination between homozygous and heterozygous resistant individuals is an advantage over other methods, such as PASA or the loop-mediated isothermal amplification method currently in use for detection of ACCase-inhibitor resistance (Burgos et al. Reference Burgos, Tranel, Streibig, Davis, Shaner, Norsworthy and Ritz2013). Also, this characteristic should allow for the analysis of composite samples composed of multiple individuals or even multiple populations, reducing the number of samples needed to identify localities or regions where the resistance mutation is present.
Normalized (Figure 5a) and difference plots (Figure 5b) showed a clear clustering of resistant and susceptible alleles in the samples. Additionally, DNA sequences of the target region (Figure 6) perfectly matched those obtained using HRMA, and no false-positive or false-negative signals were detected. These results showed that HRMA is a suitable alternative method to sequencing for detection of mutations conferring resistance to ACCase-inhibiting herbicides, which is also advantageous for possible use in geographical regions where access to sequencing is limited. Nonetheless, equipment cost is rather high, meaning that there is a substantial initial investment required and the method is likely to be applied by laboratories with good financial support (Chatzidimopoulos et al. Reference Chatzidimopoulos, Ganopoulos, Vellios, Madesis, Tsaftaris and Pappas2014).
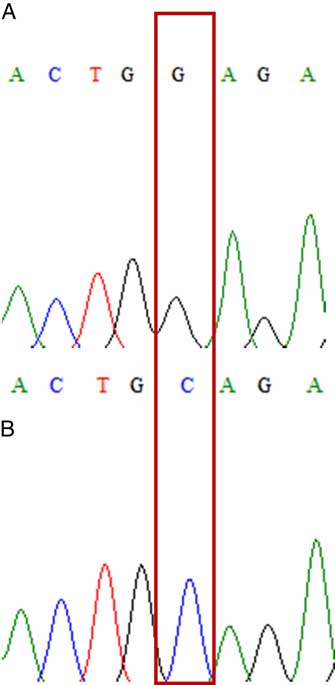
Figure 6 Direct sequence results of two genotypes of the Trp-2027-Cys (G>C) mutation in Rottboellia cochinchinensis samples. (A) Wild type and (B) mutant type.
While HRMA has many advantages over other methods for detection of ACCase mutations, there are some limitations similar to those stated for fungicide-resistance detection by Samaras et al. (Reference Samaras, Madesis and Karaoglanidis2016). The most important limitation is the need for reference controls. Reference and unknown samples should be analyzed at the same time to allow for direct comparison of the melting curves to control variations between sample runs. Nonetheless, once a standard curve for the reference is defined, unknown samples may be analyzed by comparing the unknown sample curve with the reference curve, which will allow for the comparison of results from different laboratories (Ganopoulos et al. Reference Ganopoulos, Aravanopoulos, Madesis, Pasentsis, Bosmali, Ouzounis and Tsaftaris2013; Samaras et al. Reference Samaras, Madesis and Karaoglanidis2016). Nevertheless, this procedure should be adopted with caution, as results may be influenced by several factors, such as DNA extraction procedure (quantity and quality), reaction mix composition, and instrument used (Ganopoulos et al. Reference Ganopoulos, Aravanopoulos, Madesis, Pasentsis, Bosmali, Ouzounis and Tsaftaris2013). A second limitation is that this method is qualitative and does not provide a quantitative measure like the frequency of alleles within a population (Samaras et al. Reference Samaras, Madesis and Karaoglanidis2016). Finally, it is important to take into consideration in a large-scale monitoring program the probability for the presence of natural polymorphisms, which are not necessarily implicated in resistance (Kaundun Reference Kaundun2014), in the amplified products that might change the melting profile of the amplicon, leading to false positives related to wild type. This probability is considered low for the plastidic ACCase gene due to its relatively conserved nature (Kaudun 2014). Also, keeping the length of the PCR amplicon as small as possible reduces the risk for detection of natural polymorphisms. Moreover, R. cochinchinensis plants carrying another ACCase-inhibitor resistance mutation outside the PCR fragment flanked by the primers will appear to be susceptible although they are resistant (false negative).
A strategy frequently used in mutation screening is to pool multiple samples to reduce the number of samples to be analyzed, thus increasing the efficiency of the screen, while reducing overall costs of the assay. The method we developed can be integrated into this type of monitoring system by allowing for composite samples from different regions (i.e., each sample containing multiple populations of the same region) to be processed first, and once a positive sample is detected, a second analysis of individual populations to identify the population where the resistant allele is present can be made. Finally, sequencing can be used to confirm the positive results. It has been shown that HRMA can detect mutations in samples containing from 1% to 10% of mutant DNA in the background of wild-type DNA (Liu et al. Reference Liu, Wu, Yang, Xu, Chen, Huang and Fu2014), although using composite samples may lower sensitivity, thus making it more difficult to detect mutations that are present at low frequency in a wild-type background (Li et al. Reference Li, Chu, Liu, Jing, Liu and Hao2010; Simko Reference Simko2016). Therefore, further titration studies are needed to identify the number of samples that can be combined to identify the resistant mutation in a predominantly susceptible sample.
Many ACCase herbicide–resistant grass weed species are polyploids (auto- and allopolyploids) with multiple homoeologous plastidic ACCase gene copies containing ACCase-resistant mutations (Beckie and Tardif Reference Beckie and Tardif2012; Yu et al. Reference Yu, Ahmad-Hamdani, Han, Christoffers and Powles2013). Although it was not assessed in this study, small amplicon analysis using HRMA also proved to be a sensitive and consistent method for SNP genotyping and assaying allelic dosage in polyploidy species (Han et al. Reference Han, Khu and Monteros2012; Simko Reference Simko2016).
Although we acknowledge that further evaluation should be carried out on a larger number of samples, the RT-PCR HRMA presented in this study has the potential to be used in a routine manner as a high-throughput technique for genotyping fluazifop-P-butyl resistance in R. cochinchinensis based on the C/G mutation (Trp-2027-Cys) within the amplified ACCase gene fragment. This method could be extended to other resistance-associated mutations within the ACCase gene and also to other target-site mutation herbicide-resistance mechanisms in other species.
Acknowledgments
The authors would like to thank Ramon Leon from North Carolina State University and Paul Esker from Penn State University for their input and critical review of the article.








