Infectious diseases may demonstrate a heritable component — that is, the propensity to contract and develop active infection and the severity of the immune response — and is influenced not only by contact with the infectious agent but also by host genetic factors (Chapman & Hill, Reference Chapman and Hill2012). For instance, host genetic variants mediating the immune response, including the biosynthesis of host glycan structures, are associated with viral diversity (Fumagalli et al., Reference Fumagalli, Pozzoli, Cagliani, Comi, Bresolin, Clerici and Sironi2010); immunogenetic factors are implicated in the risk and severity of influenza caused by H1N1 infection (Keynan et al., Reference Keynan, Malik and Fowke2013). Little is known about host genetic factors influencing the infection of humans by coronaviruses in general, and almost nothing is known for SARS-CoV-2 in particular. Nevertheless, a recent genome-wide association study identified two genomic regions associated with respiratory failure caused by COVID-19 infection (Severe Covid-19 GWAS Group et al., Reference Ellinghaus, Degenhardt, Bujanda, Buti, Albillos and Karlsen2020). A possible role of variation in ACE2 and TMPRSS2 genes in COVID-19 spread and severity is being explored (Delanghe et al., Reference Delanghe, Speeckaert and De Buyzere2020; Lopera Maya et al., Reference Lopera Maya, van der Graaf, Lanting, van der Geest, Fu and Swertz2020; Strope et al., Reference Strope, Pharm and Figg2020).
We developed the C-19 COVID-19 symptom tracker app, which was launched on March 24, 2020, to collect real-time data during the SARS-CoV-2 pandemic (Drew et al., Reference Drew, Nguyen, Steves, Menni, Freydin and Varsavsky2020). Once downloaded by a participant, the app asks about age, health risk factors and location. It collects information about symptoms after an initial screening question to determine whether the participant feels well or not. Participants are invited to contribute further updates every few days regarding their symptoms, healthcare visits, COVID-19 testing and results and whether they were in quarantine.
The aim of the current study was to estimate the heritability of COVID-19 symptoms in app participants who are also twins registered with the TwinsUK study and identify phenotypes suitable for genome-wide association study that would shed light on the host genetic mechanisms of infection and symptom generation.
Materials and Methods
The app asks, on a daily basis, about how the participant feels and the presence or absence of common symptoms including cough, fever, chest pain, delirium and anosmia, and has been downloaded by >3 million people. Participants in the TwinsUK adult twin register (Verdi et al., Reference Verdi, Abbasian, Bowyer, Lachance, Yarand, Christofidou and Steves2019) who had reported current symptoms via the app were included in this study.
The stem question is: ‘How do you feel now right now?’ The possible answers include: ‘I feel as healthy as normal’ and ‘I am not feeling quite right’. In case of answering ‘I am not feeling quite right’, another set of questions follows concerning COVID-19 related symptoms: fever, fatigue (mild or severe), delirium, persistent cough, shortness of breath (mild, severe, significant), anosmia (loss of smell and test), chest pain, abdominal pain, hoarse voice, diarrhea and loss of appetite (skipped meals). For the purpose of current study, individuals replying ‘I feel as healthy as normal’ have been assigned negative values (0) for all questions. For those reporting not feeling quite right, positive (1) or negative status was assigned based on the response to each question. Fatigue was considered positive if reported as severe and negative if reported as absent or mild. Shortness of breath was considered positive if reported as being severe or significant and negative if reported as absent or mild.
Casewise concordance between twin pairs and its standard error were estimated using the likelihood-based approach (Witte et al., Reference Witte, Carlin and Hopper1999). Heritability of individual symptoms and ‘predicted COVID-19’ was estimated using biometric modeling for the liability threshold model (Falconer & Mackay, Reference Falconer and Mackay1996). The liability-threshold model assumes an underlying continuous liability that follows a normal distribution. The model decomposes the variance of the liability into latent sources of variation: additive genetic (A), common environment (C) and unique environment variance (E). For each symptom, we fitted saturated model and models with the liability variance components: ACE, AE, CE and E. Models were compared using chi-squared statistic estimated as double the difference between log-likelihoods of the models, which under null follows the central chi-squared distribution with degrees of freedom equal to the difference between the number of estimated parameters for the models. The ACE model was compared with the saturated model; AE and CE models were compared with ACE and E model was compared with either AE or CE depending on which one provided the best fit. Narrow-sense heritability, the proportion of the variance in liability caused by additive genetic effects, was estimated from the most parsimonious model (with adjustment for age, sex and body mass index [BMI]) as determined by the Akaike information criterion (Akaike, Reference Akaike1974). Biometric modeling was performed using OpenMx package for R (Neale et al., Reference Neale, Hunter, Pritikin, Zahery, Brick, Kirkpatrick and Boker2016).
‘Predicted COVID-19’ status was determined by linear combination of self-reported age, sex and symptoms of anosmia, severe or significant persistent cough, fatigue and skipped meals. We have shown this model to be most predictive of SARS-CoV-2 positivity on a swab PCR test (Menni et al., Reference Menni, Valdes, Freidin, Sudre, Nguyen, Drew and Spector2020). We also examined real-time information obtained by SMS text from TwinsUK participants regarding their current cohabiting arrangement. To control for the influence of infection within households on estimates of C component, we repeated the heritability estimates excluding twins living together and those with cohabiting information missing. We also performed the estimates adjusting for the index of multiple deprivation.
Ethics committee approval was obtained for TwinsUK from St Thomas’ Hospital Ethics Committee 2008 with further approval obtained to use health records for research.
Results
Adult same-sex twins (n = 3261) had provided data for analysis between March 25 and October 13, 2020 (Table 1). The sample included 3099 individuals of White British ancestry comprising 1175 unpaired twins, 674 total pairs of monozygotic (MZ) twins and 288 pairs of dizygotic (DZ) twins that have been used to calculate heritability estimates. There were differences in mean age between unpaired twins and paired twins of both zygosities and higher BMI compared to MZ twins. There were differences between MZ and DZ twins for age and BMI. Also, consistent with expectations, DZ twins lived further apart from each other compared to MZ twins. Prevalence of symptoms in TwinsUK was similar to the larger dataset of n = 3.45 million (Figure 1).
Table 1. Demographics of the studied twins
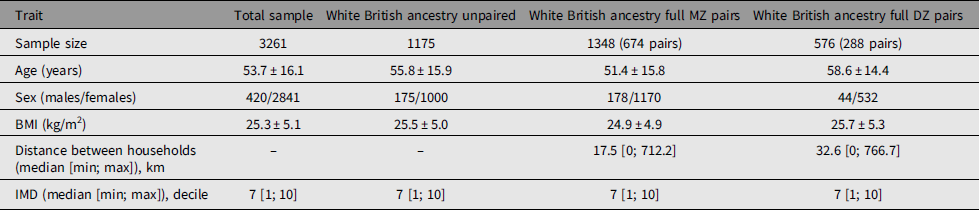
Note: BMI, body mass index; IMD, index of multiple deprivation.

Fig. 1. Prevalence of symptoms and ‘Predicted COVID-19’ status reported by TwinsUK twins and non-twin population reported the symptoms using C-19 COVID-19 symptom tracker app.
Table 2 shows prevalence of symptoms in the MZ and DZ twins and the casewise concordance and tetrachoric correlations. Based on the greater concordance and tetrachoric correlations in MZ twins compared to DZ twins, a heritable component is suggested for delirium, diarrhea, fatigue, anosmia, predicted COVID-19 and skipped meals. This was confirmed by biometric modeling, with the AE, model being the best fit for these traits and the following heritability estimates: delirium, 49% (95% CI [32, 64]); diarrhea, 34% (95% CI [20, 47]); fatigue, 31% (95% CI [8, 52]); anosmia, 19% (95% CI [0, 38]); predicted COVID-19, 31% (95% CI [11, 48]) and skipped meals, 46% (95% CI [31, 60]). For other traits, the CE was found to be the best-fitting model, suggesting a higher impact of environmental factors.
Table 2. Prevalence, casewise concordance and tetrachoric correlation of the symptoms reported in TwinsUK

Note: Only White British ancestry complete twin pairs are included. Values in bold type represent nominally statistically significant differences in estimated parameters.*SE unidentifiable due to absence of concordant cases.
Heritability estimates, including only those twin pairs living apart and adjusting for intrapair distance (Supplementary Table S1) and adjusting for index of multiple deprivation (Supplementary Table S2), were largely similar.
Discussion
Here, we report that 31% of the variance of ‘predicted COVID-19’ phenotype is due to genetic factors. This value is in line with heritability estimates for other viral infections: the heritability of HIV acquisition is estimated to be 28−42% (Powell et al., Reference Powell, Duarte, Hotopf, Hatch, de Mulder Rougvie, Breen and Nixon2020) and the heritability of hepatitis B viral load is 30−70% (Huang et al., Reference Huang, Shih, Li, Wu, Chen, Lin and Yu2011). We found that delirium, a symptom related to immune activation, has a high heritability (49%). The symptom of anosmia, previously reported by us to be an important predictive symptom of COVID-19, was also heritable at 19%. Anosmia is now considered a key and a rather specific symptom of COVID-19 at the early stage of the infection (Gengler et al., Reference Gengler, Wang, Speth and Sedaghat2020; Passarelli et al., Reference Passarelli, Lopez, Mastandrea Bonaviri, Garcia-Godoy and D’Addona2020). While the precise pathophysiology of anosmia associated with COVID-19 is not yet fully understood, it may be related to sino-nasal viral shedding, important as a key virus transmission mechanism (Gengler et al., Reference Gengler, Wang, Speth and Sedaghat2020). That heritable factors influence anosmia may help identify genetic variants responsible for susceptibility to infection. Symptomatic infection with SARS-CoV-2, rather than representing a purely stochastic event, is under host genetic influence to some extent and may reflect inter-individual variation to viral infection susceptibility and in the host immune response. Viral infections typically lead to T-cell activation with IL-1, IL-6 and TNF-α release causing flu-like symptoms such as fever. The genetic basis of this variability in response will provide important clues for therapeutics and lead to the identification of groups at high risk of death, which is associated with a cytokine storm approximately 2 weeks after symptom onset (Vaninov, Reference Vaninov2020).
Interestingly, for feeling unhealthy, abdominal pain, chest pain, hoarse voice, persistent cough and shortness of breath, we found the common environment to be more pronounced than the heritable component. Our sensitivity analysis including only twins living apart confirmed this (Supplementary Table S1). Another sensitivity analysis with adjustment for the possible social disparity was also in keeping with this observation (Supplementary Table S2). Importantly, the majority of these traits concern the airways, suggesting that they may have been exaggerated by the winter/early spring weather conditions inadvertently shared by most of the twins regardless of their living place.
Among unanticipated findings, we highlight a heritable component for skipped meals and diarrhea (Table 3) in keeping with evidence for active replication of SARS-CoV-2 in the digestive tract, even after clearance of the virus from the respiratory tract, and a high level of gastrointestinal symptoms in affected patients (Yang & Tu, Reference Yang and Tu2020). This implies that host genetic factors control the manifestation of COVID-19 infection in the gut, perhaps by regulation of inflammation; an interplay with gut microbiota, which in turn is heritable, could also account for our findings (Zuo et al., Reference Zuo, Zhang, Lui, Yeoh, Li, Zhan and Ng2020).
Table 3. Biometric modeling for COVID-19 symptoms in TwinsUK
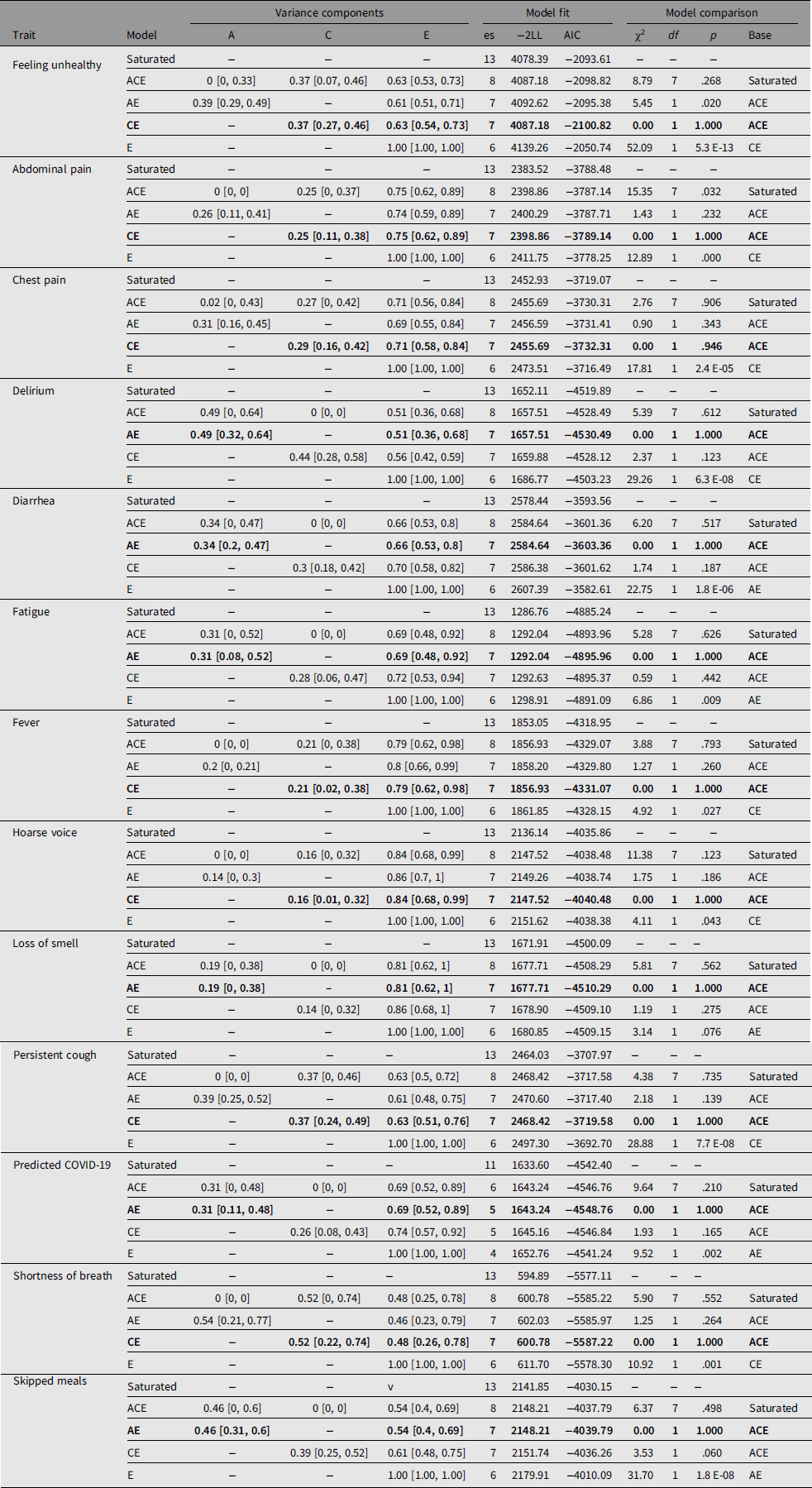
Note: Biometric modeling was carried out using OpenMx package for R as detailed in the main text. Adjustments were done for age, sex and BMI for all traits except predicted COVID-19 for which no adjustment for age and sex was done. Bold type indicates the best-fit models according to AIC and χ2 tests. 95% confidence intervals are shown in square brackets. es, number of estimated parameters in the model; −2LL, log-likelihood for the model multiplied by −2; AIC, Akaike Information Criteria; χ2, test statistics for models comparison; df, degrees of freedom for χ2; p value, corresponding p value; base, model against which the comparison was done.
The study has a number of limitations. First, our data are likely to suffer from a healthy volunteer bias. Indeed, we report negative tetrachoric correlations for DZ twins for anosmia and predicted COVID-19 due to the few pairs concordant for rare symptoms. However, all methods currently available — testing healthcare workers, testing those admitted to hospital with symptoms, testing those with symptoms at home — have limitations that lead to inherent bias. Ours is among the best representations of the general population (Drew et al., Reference Drew, Nguyen, Steves, Menni, Freydin and Varsavsky2020), albeit at the healthy end of the spectrum, and it offers an important contribution to the accurate depiction of the totality of the epidemic. Another limitation is the lack of information about the exposure of the twins to SARS-CoV2. One can infer that COVID-19 cases are more likely to have been exposed to the virus than controls. This may be true for healthcare and social workers on the frontline of COVID-19 treatment and management. However, this does not necessarily apply to the general population, as a large proportion of people (40−45%) are reported to be asymptomatic (Oran & Topol, Reference Oran and Topol2020), so cases cannot fully resemble exposure rates. Nevertheless, there is nothing to suggest that differential levels of virus exposure between MZ and DZ twins could have impacted heritability estimates. Another limitation is that many symptoms are nonspecific and are prevalent in spring in the Northern hemisphere when allergies and seasonal influenza are active, and are not indicative of infection status. In line with that, as a large proportion of twins logged their symptoms only once, we cannot assess the consistency of reporting for them. Among those who logged more than once, the following are prevalences of reporting the presence of symptoms more than once: feeling unhealthy, 71.4%; persistent cough, 64.4%; loss of smell, 62.7%; hoarse voice, 55.5%; shortness of breath, 51.8%; chest pain, 51.8%; delirium, 47.1%; skipped meals, 46.9%; fatigue, 42.1%; fever, 41.5%; abdominal pain, 37.8% and diarrhea, 36.8%. This may reflect both the course of the disease or changes in disease status perception in the responders, with the latter being a possible bias. We did have a sufficient sample size of people reporting symptoms and viral test results to indicate accurately which symptom pattern provides the greatest positive predictive value. The study could have been biased by MZ twins cohabiting more than DZ twins, but real-time data collection allowed us to exclude cohabiting pairs. Also, adjusting for index of multiple deprivation, we controlled for the possible disparity between different regions of the country. Our twin sample is predominantly female for historical recruitment reasons and is not representative of non-European ancestries (Verdi et al., Reference Verdi, Abbasian, Bowyer, Lachance, Yarand, Christofidou and Steves2019). Finally, concerning predicted COVID-19, the predictive model is based on the results of RT-PCR tests, which themselves are not perfect; and the model itself is not 100% sensitive and specific. Nevertheless, RT-PCR test results are well correlated with chest CT-scan for the diagnosis of COVID-19 (Ai et al., Reference Ai, Yang, Hou, Zhan, Chen, Lv and Xia2020). Also, the model we used to predict COVID-19 has been validated in an independent dataset in the USA (Menni et al., Reference Menni, Valdes, Freidin, Sudre, Nguyen, Drew and Spector2020), suggesting that it has utility. Finally, there is a fair concordance between predicted COVID-19 and positive COVID-19 antibodies in the studied twins with Yule’s coefficient of colligation of .52 (.46−.57) and Cohen’s kappa of .36 (.29−.42).
Our study has several strengths. TwinsUK participants and their symptom reporting is representative of the UK population (Figure 1). Predicting infection with a validated combination of symptoms of virus-tested individuals offers a pragmatic solution to the challenges of widespread testing in the general population. One of the important contributions of our study is the real-time nature of the data collection and the very large sample drawn from the general population (Drew et al., Reference Drew, Nguyen, Steves, Menni, Freydin and Varsavsky2020). While acknowledging the wide confidence intervals around the estimates of heritability, our study contributes to the understanding of COVID-19 within the general population.
The genetic influence on COVID-19 symptoms may reflect the genotype status of candidate genes such as ACE2R, which encodes the target for viral attachment (Hoffmann et al., Reference Hoffmann, Kleine-Weber, Schroeder, Kruger, Herrler, Erichsen and Pohlmann2020), and a number of other genes, including those identified in genome-wide association studies (Elhabyan et al., Reference Elhabyan, Elyaacoub, Sanad, Abukhadra, Elhabyan and Dinu2020). Another interesting possibility concerns the genetic factors underlying psychological and behavioral traits. Anxiety, depression, stress and disturbed sleep have been reported to be common psychological reactions to the COVID-19 pandemic (Rajkumar, Reference Rajkumar2020). Hereditable factors play a role in the variation of these traits (Smoller, Reference Smoller2016), and therefore may contribute to COVID-19 symptom reporting. Further genetic work is underway to determine whether twin genotype at ACE2R influences predicted positivity or symptoms and a global genetic study is underway (COVID-19 Host Genetics Initiative, 2020). Public health approaches to identify those at increased genetic risk of severe infection would be useful as a way of partially mitigating the economic effects of lockdown and social distancing policies.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/thg.2020.85.
Acknowledgments
We are grateful to all the users of the C-19 COVID-19 symptom tracker app for their contribution to this study.
Financial Support
FW receives funding for COVID-19 research from the Kennedy Trust and Versus Arthritis. TwinsUK is funded by the Wellcome Trust, Medical Research Council, European Union, Chronic Disease Research Foundation (CDRF), Zoe Global Ltd and the National Institute for Health Research (NIHR)-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London.
Conflict of Interest
T.D.S. is a consultant to Zoe Global. RD is an employee of Zoe Global. Other authors declare no conflict of interest.
Ethical Standards
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008.






