Schizophrenia and major depressive disorder (MDD) are severe mental disorders with relatively high morbidity and heritability. Schizophrenia affects about 1% of the world population and has approximately 60–85% individual heritability. Reference Burmeister, McInnis and Zöllner1 MDD affects 8–12% of the population in most countries and has an individual heritability of 40–50%. Reference O'Donovan, Craddock and Owen2 The aetiology of these two psychiatric disorders is still not clear, although genetic factors appear to be possible risk factors. Reference Burmeister, McInnis and Zöllner1 Several genetic studies have been carried out to search for the possible risk genes of these disorders. Identifying DNA variants associated with schizophrenia and MDD is a crucial step in understanding the pathophysiology of these disorders.
Transcription factor Sp4 (SP4) is a member of the SP1 family of transcription factors, which is highly expressed in the brain and could reasonably play an important role by controlling the transcription of various genes. Reference Zhou, Qyang, Kelsoe, Masliah and Geyer3,Reference Ramos, Gaudillière, Bonni and Gill4 Previously, SP4 single nucleotide polymorphisms (SNPs) have been found to be associated with schizophrenia, MDD and bipolar disorder in the White population. Reference Shi, Potash, Knowles, Weissman, Coryell and Scheftner5–Reference Zhou, Tang, Greenwood, Guo, He and Geyer7 Rare copy number variations and deletions in the SP4 gene have also been found to be associated with schizophrenia. Reference Tam, van de Lagemaat, Redon, Strathdee, Croning and Malloy8,Reference Zhou, Nie, Roberts, Zhang, Sebat and Malhotra9 Furthermore, SP4 has been found to play an important role in the development of cerebellar granule neurons by promoting activity-dependent pruning of dendritic processes. Reference Ramos, Gaudillière, Bonni and Gill4 Recently, using genome-wide association studies (GWAS) and a meta-analysis, Shyn et al Reference Shyn, Shi, Kraft, Potash, Knowles and Weissman6 reported that SP4 was a leading susceptibility candidate gene for MDD, and that it might also be the shared risk factor for both schizophrenia and bipolar disorder. Reference Blackwood, Fordyce, Walker, St Clair, Porteous and Muir10,Reference Millar, Christie, Anderson, Lawson, Hsiao-Wei Loh and Devon11 Fusté et al Reference Fusté, Pinacho, Meléndez-Pérez, Villalmanzo, Villalta-Gil and Haro12 showed that protein levels of SP1 and SP4 were significantly reduced in first-episode psychosis in lymphocytes, suggesting that these transcription factors were potential peripheral biomarkers of psychotic spectrum disorders in the early stages of the disorder.
Zhou et al Reference Zhou, Long, Geyer, Masliah, Kelsoe and Wynshaw-Boris13 showed that reduced Sp4 expression in mice induced behavioural defects associated with psychiatric disorders. Hypomorphic Sp4 mutant mice displayed robust deficits in both sensorimotor gating and contextual memory. This revealed a novel Sp4 pathway that is essential for hippocampal integrity and modulates behavioural processes.
We hypothesised that the SP4 gene might contribute to the pathogenesis of schizophrenia or MDD by modulating sensorimotor gating. We carried out our association study in the Han Chinese population. We comprehensively genotyped nine SNPs – rs1018954, rs10233357, rs2073534, rs2107448, rs2285941, rs3735440, rs40245, rs10255890 and rs6461563 – among 1235 patients with schizophrenia, 1045 patients with MDD and 1235 normal controls in the Han Chinese population.
Method
Patients
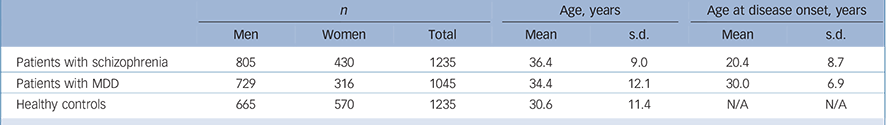
The sample set included 1235 unrelated patients with schizophrenia (805 men and 430 women), 1045 unrelated patients with MDD (729 men and 316 women) and 1235 healthy controls (665 men and 570 women). All these samples were recruited from the Han Chinese population.
Patients were recruited from the Shanghai Mental Health Center. The mean age of patients with schizophrenia was 36.4 years (s.d. = 9.0), and age at disease onset was 20.4 years (s.d. = 8.7); the mean age of patients with MDD was 34.4 years (s.d. = 12.1), and age at disease onset was 30.0 years (s.d. = 6.9); the mean age of the healthy controls was 30.6 years (s.d. = 11.4) (Table 1). Two independent psychiatrists from the Shanghai Mental Health Center interviewed each patient. Diagnoses were made strictly according to the DSM-IV criteria based on the Structured Clinical Interview for DSM-IV Axis I Disorders (SCID-I). Reference First, Spitzer, Gibbon and Williams14 Patients with the following exclusion criteria were not recruited: history of substance use; neurological illness; mental retardation; mood disorder; psychotic disorder due to general medical condition. Healthy controls were randomly selected from the general Han Chinese population and were recruited from the local community residents by means of announcements on bulletin boards. Before collecting blood samples, the healthy controls were also interviewed by two independent psychiatrists using the SCID-I to exclude any lifetime psychiatric disorders. The healthy controls were excluded only if they had a severe medical illness. Written informed consent was obtained from all participants. The study proposal and protocol were reviewed and approved by the local Ethical Committee of Human Genetics.
Table 1 Characteristics of the study sample set
| n | Age, years | Age at disease onset, years | |||||
|---|---|---|---|---|---|---|---|
| Men | Women | Total | Mean | s.d. | Mean | s.d. | |
| Patients with schizophrenia | 805 | 430 | 1235 | 36.4 | 9.0 | 20.4 | 8.7 |
| Patients with MDD | 729 | 316 | 1045 | 34.4 | 12.1 | 30.0 | 6.9 |
| Healthy controls | 665 | 570 | 1235 | 30.6 | 11.4 | N/A | N/A |
MDD, major depressive disorder; N/A, not applicable.
Selection and genotyping of SNPs
The SP4 gene is located on chromosome 7p15; it spans about 86.79 kb of DNA and consists of six exons and five introns. It contains several SNPs as identified by the International SNP Consortium (www.ncbi.nlm.nih.gov/SNP), the Ensembl genome browser (www.ensembl.org) and the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway). We selected the common tag SNPs from the HapMap (http://hapmap.ncbi.nlm.nih.gov/) Han Chinese population in Beijing. 15 The relationship between tag SNPs and the SP4 gene is shown in online Fig. DS1.
In accordance with the study protocol, the QuickGene DNA whole blood kit L (FujiFilm) was used to extract genomic DNA from peripheral blood samples. Tag SNP selection was performed with the Haploview software (Broad Institute; www.broadinstitute.org), with r 2⩾0.5 and minor allele frequency ⩾0.05. Reference de Bakker, Yelensky, Pe'er, Gabriel, Daly and Altshuler16,Reference de Bakker, Burtt, Graham, Guiducci, Yelensky and Drake17 Nine SNPs (rs1018954, rs10233357, rs2073534, rs2107448, rs2285941, rs3735440, rs40245, rs10255890 and rs6461563) were selected according to previous reports and the results of tag SNPs selection. Reference Zhou, Tang, Greenwood, Guo, He and Geyer7,Reference de Bakker, Yelensky, Pe'er, Gabriel, Daly and Altshuler16 Coverage of the whole gene by those tag SNPs was 72%, as analysed by the Tagger software (www.broadinstitute.org/mpg/tagger/server.html). The location of the nine SNPs in the studied region is shown in online Table DS1. All SNPs were genotyped with the TaqMan SNP Genotyping Assays and the Fluidigm EP1 platform. All probes and primers were designed using the Assay by Design or Assay on Demand service of Life Technologies. SNPs were determined by the genotype calls of each sample with a call rate <95%.
Statistical analysis
Association analysis with regard to the SNPs was carried out with the SHEsis software (http://analysis.bio-x.cn Reference Shi and He18,Reference Li, Zhang, He, Tang, Li and Zeng19 ), a user-friendly platform equipped with a suite of highly efficient analytical tools designed for association studies that can analyse linkage disequilibrium, haplotype construction and genetic association at polymorphism loci. SHEsis was integrated with the partitionligation-combination-subdivision expectation maximisation algorithm, designed for efficient estimation of haplotypes constructed from large numbers of biallelic or multi-allelic loci in diploid individuals. All tests were two-tailed, with statistical significance set at P<0.05. P-values were adjusted using the Bonferroni correction method.
Population stratification analysis
We used the STRUCTURE software, version 2.3.4 (http://pritchardlab.stanford.edu/structure.html Reference Falush, Stephens and Pritchard20–Reference Hubisz, Falush, Stephens and Pritchard22 ) to perform the population stratification analysis. Additionally, we genotyped the data of 79 randomly selected SNPs for the stratification analysis. We obtained the data for these 79 SNPs from 522 HapMap samples, including 174 samples from Utah residents with ancestry from Northern and Western Europe in the United States (CEU), 209 samples from Yoruba, in Ibadan, Nigeria (YRI) and 139 samples from the Han Chinese population in Beijing (CHB) (International HapMap Project; http://hapmap.ncbi.nlm.nih.gov/cgi-perl/gbrowse/hapmap28_B36/). 15 We used the STRUCTURE software to isolate the distinct populations using the genotype data of these 79 SNPs from the 522 HapMap samples. We applied the admixture and correlated frequencies models, with a burn-in length of 10 000 and Markov chain Monte Carlo repeats of 10 000.
Results
Linkage disequilibrium
The pairwise linkage disequilibrium among the nine investigated SNPs was different in the different sample sets. SNPs with D′>0.95 in both disorder sample sets were categorised in the same block. No specific haplotype block was identified, as shown in Fig. 1. (See also online Figs DS3 and DS4.)

Fig. 1 Linkage disequilibrium plot of the nine single nucleotide polymorphisms in the samples associated with (a) schizophrenia and (b) major depressive disorder.
Single site association analysis
The rs2107448 SNP was excluded from further analysis because it was not in Hardy–Weinberg equilibrium in the healthy controls, the significance threshold being P<0.05.
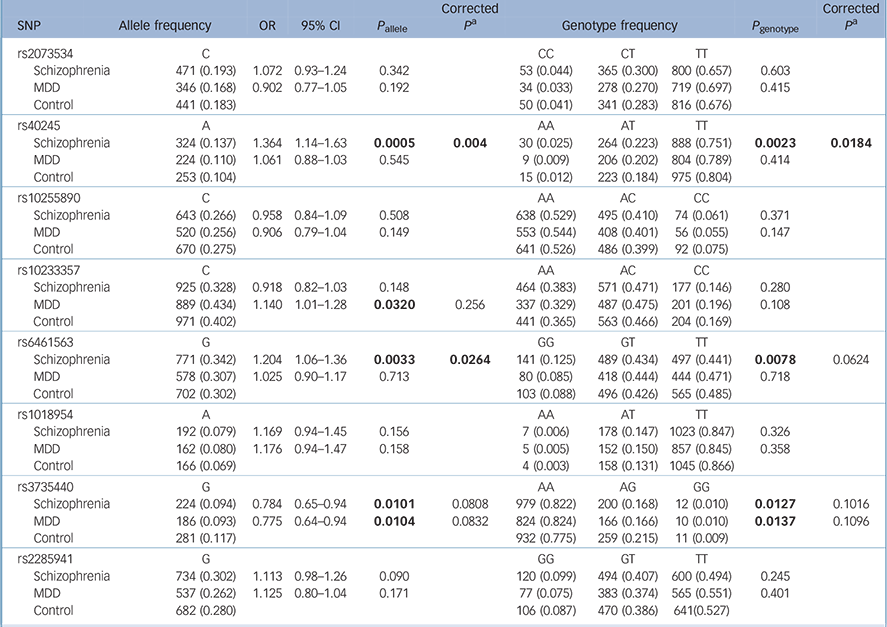
Table 2 shows the results for the remaining eight polymorphic SNPs: rs40245 was significantly associated with schizophrenia in both allele and genotype distributions (P allele = 0.0005, P allele = 0.004 after Bonferroni correction; P genotype = 0.0023, P genotype = 0.0184 after Bonferroni correction); rs6461563 was significantly associated with schizophrenia in the allele distributions (P allele = 0.0033, P allele = 0.0264 after Bonferroni correction; P genotype = 0.0078, P genotype = 0.0624 after Bonferroni correction); rs3735440 showed allelic or genotypic significance with both schizophrenia and MDD before Bonferroni correction, and rs10233357 showed allelic significance with MDD before Bonferroni correction, but these association were eliminated after Bonferroni correction.
Table 2 Results from the statistical analysis of eight single-nucleotide polymorphisms
| SNP | Allele frequency | OR | 95% CI | P allele | Corrected P a |
Genotype frequency | P genotype | Corrected P a |
||
|---|---|---|---|---|---|---|---|---|---|---|
| rs2073534 | C | CC | CT | TT | ||||||
| Schizophrenia | 471 (0.193) | 1.072 | 0.93–1.24 | 0.342 | 53 (0.044) | 365 (0.300) | 800 (0.657) | 0.603 | ||
| MDD | 346 (0.168) | 0.902 | 0.77–1.05 | 0.192 | 34 (0.033) | 278 (0.270) | 719 (0.697) | 0.415 | ||
| Control | 441 (0.183) | 50 (0.041) | 341 (0.283) | 816 (0.676) | ||||||
| rs40245 | A | AA | AT | TT | ||||||
| Schizophrenia | 324 (0.137) | 1.364 | 1.14–1.63 | 0.0005 | 0.004 | 30 (0.025) | 264 (0.223) | 888 (0.751) | 0.0023 | 0.0184 |
| MDD | 224 (0.110) | 1.061 | 0.88–1.03 | 0.545 | 9 (0.009) | 206 (0.202) | 804 (0.789) | 0.414 | ||
| Control | 253 (0.104) | 15 (0.012) | 223 (0.184) | 975 (0.804) | ||||||
| rs10255890 | C | AA | AC | CC | ||||||
| Schizophrenia | 643 (0.266) | 0.958 | 0.84–1.09 | 0.508 | 638 (0.529) | 495 (0.410) | 74 (0.061) | 0.371 | ||
| MDD | 520 (0.256) | 0.906 | 0.79–1.04 | 0.149 | 553 (0.544) | 408 (0.401) | 56 (0.055) | 0.147 | ||
| Control | 670 (0.275) | 641 (0.526) | 486 (0.399) | 92 (0.075) | ||||||
| rs10233357 | C | AA | AC | CC | ||||||
| Schizophrenia | 925 (0.328) | 0.918 | 0.82–1.03 | 0.148 | 464 (0.383) | 571 (0.471) | 177 (0.146) | 0.280 | ||
| MDD | 889 (0.434) | 1.140 | 1.01–1.28 | 0.0320 | 0.256 | 337 (0.329) | 487 (0.475) | 201 (0.196) | 0.108 | |
| Control | 971 (0.402) | 441 (0.365) | 563 (0.466) | 204 (0.169) | ||||||
| rs6461563 | G | GG | GT | TT | ||||||
| Schizophrenia | 771 (0.342) | 1.204 | 1.06–1.36 | 0.0033 | 0.0264 | 141 (0.125) | 489 (0.434) | 497 (0.441) | 0.0078 | 0.0624 |
| MDD | 578 (0.307) | 1.025 | 0.90–1.17 | 0.713 | 80 (0.085) | 418 (0.444) | 444 (0.471) | 0.718 | ||
| Control | 702 (0.302) | 103 (0.088) | 496 (0.426) | 565 (0.485) | ||||||
| rs1018954 | A | AA | AT | TT | ||||||
| Schizophrenia | 192 (0.079) | 1.169 | 0.94–1.45 | 0.156 | 7 (0.006) | 178 (0.147) | 1023 (0.847) | 0.326 | ||
| MDD | 162 (0.080) | 1.176 | 0.94–1.47 | 0.158 | 5 (0.005) | 152 (0.150) | 857 (0.845) | 0.358 | ||
| Control | 166 (0.069) | 4 (0.003) | 158 (0.131) | 1045 (0.866) | ||||||
| rs3735440 | G | AA | AG | GG | ||||||
| Schizophrenia | 224 (0.094) | 0.784 | 0.65–0.94 | 0.0101 | 0.0808 | 979 (0.822) | 200 (0.168) | 12 (0.010) | 0.0127 | 0.1016 |
| MDD | 186 (0.093) | 0.775 | 0.64–0.94 | 0.0104 | 0.0832 | 824 (0.824) | 166 (0.166) | 10 (0.010) | 0.0137 | 0.1096 |
| Control | 281 (0.117) | 932 (0.775) | 259 (0.215) | 11 (0.009) | ||||||
| rs2285941 | G | GG | GT | TT | ||||||
| Schizophrenia | 734 (0.302) | 1.113 | 0.98–1.26 | 0.090 | 120 (0.099) | 494 (0.407) | 600 (0.494) | 0.245 | ||
| MDD | 537 (0.262) | 1.125 | 0.80–1.04 | 0.171 | 77 (0.075) | 383 (0.374) | 565 (0.551) | 0.401 | ||
| Control | 682 (0.280) | 106 (0.087) | 470 (0.386) | 641(0.527) | ||||||
CI, confidence interval; MDD, major depressive disorder; OR, odds ratio; SNP, single nucleotide polymorphism.
a. Bonferroni correction.
Separate analyses for men and women are shown in online Tables DS2 and DS3. In the male samples, rs40245 was significantly associated with schizophrenia in both allele and genotype distributions, and rs6461563 was significantly associated with schizophrenia in the allele distributions.
Population stratification analysis
Online Fig. DS2 shows the population stratification analysis when K = 3: each angle represents a possible independent ancestry and the differently coloured dots represent the individuals in the population we want to analyse. The combined populations of CEU, YRI and our samples display a clearly stratified pattern (Fig. DS2(a)), and our samples of the two disorders and healthy controls are evenly distributed in the triangle indicating that there is no obvious significant stratification in the population (Fig. DS2(b)). The results were consistent with one another when K ranged from 2 to 5. We could therefore conclude that our positive results were unlikely to be caused by population stratification.
Discussion
Psychiatric disorders are likely to be caused by multiple susceptibility genes, each with small effects in increasing the risk of illness. Reference Cannon and Keller23 In this study, we discovered an association of SP4 variants with schizophrenia. Our main finding was a significant association between the SP4 gene and schizophrenia in the Han Chinese population. We confirmed that two SNPs, rs40245 (P allele = 0.0005, odds ratio (OR) = 1.364, 95% confidence interval (CI) 1.14–1.63) and rs6461563 (P allele = 0.0033, OR = 1.204, 95% CI 1.06–1.36), were associated with schizophrenia in a case–control study with large sample sizes of a Han Chinese origin. We also found that rs3735440 showed allelic and genotypic significance with both schizophrenia and MDD before Bonferroni correction, whereas rs10233357 showed allelic significance with MDD before Bonferroni correction. However, the association was too weak and they were eliminated after Bonferroni correction. We found that rs40245 and rs6461563 were more sensitive in men with schizophrenia than in women. However, the OR was in the same direction between the two genders. The female sample size was 430, which might not be large enough to detect the differences. Sample size might be one of the main reasons for the negative results. However, possible gender differences could not be excluded, and a larger female sample size is needed to reach sufficient statistical power.
SP4, located on chromosome 7p15 (start: 21,467,652, end: 21,554,440), encodes the brain-specific, zinc finger transcription factor Sp4. Sp4 recognises GC-rich sequences in the promoters of a variety of genes, Reference Heisler, Torti, Boutros, Watson, Chan and Winegarden24 where a susceptibility locus was suggested for a broad spectrum of human mental disorders. Zhou et al Reference Zhou, Tang, Greenwood, Guo, He and Geyer7 genotyped 506 patients with bipolar disorder and 507 controls, and established that four SNPs in SP4 (rs40245, rs12673091, rs1018954 and rs3735440) displayed significant association with bipolar disorder. We recruited more case and control samples in our study, and found that SNP rs40245 was a susceptibility locus for schizophrenia. At the same time, we found that rs3735440 was significantly associated with schizophrenia and MDD before Bonferroni correction, and rs10233357 was associated with MDD before Bonferroni correction. Evidence from clinical, epidemiological and molecular genetic studies has suggested that some genetic risk factors are shared between neuropsychiatric disorders. Our study also suggested that there might be overlap of genetic factors in major psychiatric disorders. Common variants nominally showed cross-disorder effects for the adult-onset disorders (schizophrenia and MDD). These studies provided direct evidence for SP4 as a susceptibility gene for mental disorders, which implied that SP4 might have some important function in the biological processes of neuronal development in humans. SP1 and SP4 transcription factors regulate gene expression in many biological processes including neuronal development and function. Reference Bouwman and Philipsen25,Reference Black, Black and Azizkhan-Clifford26 In contrast to the ubiquitous expression pattern of the SP1 gene, SP4 is broadly expressed in neurons of the central nervous system and it shows higher expression in the cerebellum and hippocampus. Reference Zhou, Long, Geyer, Masliah, Kelsoe and Wynshaw-Boris13,Reference Supp, Witte, Branford, Smith and Potter27–Reference Zhu, Nguyen, Brown, Pourhosseini, Garcia and van Bilsen29 In patients with schizophrenia, it was reported that SP1 mRNA levels altered in lymphocytes and some areas of the brain. Reference Ben-Shachar and Karry30
In addition, SP4 expression was reduced in the prefrontal cortex and cerebellum of patients with bipolar disorder, providing evidence that this gene might be relevant in psychiatric diseases with psychotic features. Reference Pinacho, Villalmanzo, Roca, Iniesta, Monje and Haro31 Recently, Fusté et al Reference Fusté, Pinacho, Meléndez-Pérez, Villalmanzo, Villalta-Gil and Haro12 assessed protein and gene expression levels of SP1 and SP4 in peripheral blood mononuclear cells of patients with first-episode psychosis and in healthy controls. They found that SP1 and SP4 levels were significantly reduced in patients compared with healthy controls, suggesting that these transcription factors might be potential peripheral biomarkers of psychotic spectrum disorders in the early stages. Shyn et al Reference Shyn, Shi, Kraft, Potash, Knowles and Weissman6 carried out a meta-analysis of three European-ancestry MDD GWAS data-sets: the Sequenced Treatment Alternatives to Relieve Depression (STAR*D) collaborative study, the Genetics of Recurrent Early-Onset Depression (GenRED) project and the publicly available Genetic Association Information Network-MDD data-set. An intronic SNP (rs17144465) in SP4 was observed for association in the meta-analysis. In our study two SNPs, rs3735440 and rs10233357, were found to be associated with MDD before Bonferroni correction, which might imply a nominal association with MDD. However, there was no association after Bonferroni correction.
So far, many functional studies have been conducted regarding the expression of SP4. The Sp4 protein is degraded with the activation of glutamate receptors. Reference Mao, Yang, Simpkins and Barger32 Reduced expression of the SP4 gene in mice exhibited decreased granule cell density in the dentate gyrus of the hippocampus, Reference Zhou, Qyang, Kelsoe, Masliah and Geyer3 deficits in sensorimotor gating and contextual learning, Reference Zhou, Long, Geyer, Masliah, Kelsoe and Wynshaw-Boris13 suggesting a possible behavioural deficit, Reference Supp, Witte, Branford, Smith and Potter27 and association with schizophrenia and other psychiatric disorders. Reference Zhou, Long, Geyer, Masliah, Kelsoe and Wynshaw-Boris13,Reference Braff, Geyer and Swerdlow33–Reference Daumas, Halley and Lassalle37 SP4 may also play a role in glutamate-induced neurotoxicity. Reference Mao, Yang, Simpkins and Barger32,Reference Mao, Moerman-Herzog, Wang and Barger38 An imbalance in dopamine and glutamate pathways may induce the emergence of these psychiatric disorders. All this evidence strongly suggests a key role for SP4 in mental disorders. Although the two risk polymorphisms rs40245 and rs6461563 are both located in the intron region, there may be some association with other functional variants. SNPs in non-coding areas may also affect gene splicing, transcription factor binding and mRNA degradation.
Our results suggest an association between SP4 and schizophrenia, although not between SP4 and MDD, in the Han Chinese population. We have shown that the same genetic factors can be shared by different mental disorders. Although our sample size was relatively large and we analysed a considerable sample size and achieved association significance of common variants, we lack further evidence from functional experiments to support our findings and to show the role of SP4 or related pathways in the pathogenesis of mental disorders.
Funding
This work was supported by the National Natural Science Foundation of China (31325014, 81130022, 81171271, 81272302, 31000553, 81121001, 31240001, 31271140), the National 863 project (2012AA02A515, 2012AA021802), the National Basic Research Program (973 Program, 2015CB559100), the Program for Changjiang Scholars and Innovative Research Team in University (IRT1025), the Foundation for the Author of National Excellent Doctoral Dissertation of China (201026), the Shanghai Rising-Star Program (12QA1401900), the Shanghai Municipal Natural Science Foundation (11ZR1431300), the Shanghai Key Laboratory of Psychotic Disorders (13dz2260500), the project of the Shanghai Mental Health Center (2012-YJ-08), the Youth Research Project of Shanghai Health and Family Planning Commission (20134Y082), Shu Guang project supported by Shanghai Municipal Education Commission and Shanghai Education Development Foundation (12SG17), and the Shanghai Jiao Tong University Liberal Arts and Sciences Cross-Disciplinary Project (13JCRZ02).
Acknowledgements
We are deeply grateful to all who took part in this study, and to the psychiatrists for their help in recruiting patients and diagnosing schizophrenia and MDD in our patient cohort.






eLetters
No eLetters have been published for this article.