The prevalence of major depressive disorder (MDD) among women is 1.5 to 3 times higher than in men.Reference Kessler, Berglund, Demler, Jin, Koretz and Merikangas1 In particular, women show heightened vulnerability for mood disorders in hormonal transition phases such as during puberty, pre- to postpartumReference Munk-Olsen, Laursen, Pedersen, Mors and Mortensen2 and perimenopause.Reference Freeman, Sammel, Boorman and Zhang3 Perinatal depression (PND) is a common, debilitating disorder with a prevalence of 19%Reference Gavin, Gaynes, Lohr, Meltzer-Brody, Gartlehner and Swinson4 that confers long-term detrimental effects on the health of both mother and child, which include adverse effects on mother–infant bonding and infant socio-cognitive development. Several biological markers, including sex-steroid hormonal fluctuations and alterations in the serotonergic system,Reference Barth, Villringer and Sacher5, Reference Frokjaer, Pinborg, Holst, Overgaard, Henningsson and Heede6 have been implicated in the pathophysiology of PND. In particular, oestrogen signalling affects the brain via diverse molecular mechanisms, including effects on hippocampal neurogenesis, neurotrophin signalling and stress hormone-axis function.Reference Krishnan and Nestler7 Oestrogen receptors are mainly nuclear receptors that serve as transcription factors by regulating the expression of target genes.Reference O'Malley8 Upon activation, the oestrogen receptors bind to DNA-enhancer elements called oestrogen-response elements, which allow the oestrogen receptors to influence transcription of their target genes. Further, oestrogen can induce local changes in DNA methylation, effects which may be driven via the oestrogen receptors. Enhanced sensitivity to oestrogen signalling, rather than abnormal absolute levels of steroid hormones, may drive increased risk for depressive symptoms when exposed to sex-steroid hormone fluctuations across peripartum.Reference Bloch, Schmidt, Danaceau, Murphy, Nieman and Rubinow9, Reference Guintivano, Arad, Gould, Payne and Kaminsky10 We previously identified 116 genes whose expression levels in the highly sex steroid-stimulated state of the third trimester of pregnancy predicted later depression with postpartum onset. These predictive transcripts were significantly enriched for oestrogen-receptor targets and showed an increased dynamic change over pregnancy and the postpartum period in women with depression at postpartum onset, suggesting an enhanced sensitivity to oestrogen signalling in this group.Reference Mehta, Newport, Frishman, Kraus, Rex-Haffner and Ritchie11 In another recent study, we used a gonadotropin-releasing hormone agonist (GnRHa) to induce a biphasic fluctuation in ovarian sex hormones as a pharmacological risk model for hormone-induced mood changes. We evaluated whether GnRHa treatment triggers depressive symptoms in a manner dependent on net changes in oestradiol levels and brain serotonin-transporter (SERT) binding.Reference Frokjaer, Pinborg, Holst, Overgaard, Henningsson and Heede6 Treatment with GnRHa induces a biphasic hormonal transition phase reflected by an initial stimulation of the hypothalamic–pituitary–gonadal axis; a desensitisation of the pituitary GnRH receptors; and subsequently a suppression of the ovarian sex-steroid production to menopausal level, which is reached within 10–14 days and sustained for 28 days post-intervention. Using this pharmacologically induced, biphasic, ovarian-hormone response by GnRHa via a randomised, double-blinded, placebo-controlled, within-subject study design, we have observed a significant association between SERT brain binding (measured with [11C]DASB Positron Emission Tomography (PET)) and scores from the 17-item Hamilton Rating Scale for Depression (HRSD-17) following GnRHa treatment.Reference Frokjaer, Pinborg, Holst, Overgaard, Henningsson and Heede6 This suggests that serotonergic signalling and oestradiol changes were key to mechanisms in hormone fluctuation-provoked depressive symptoms.
In this study, we have combined our findings from a previous study on biomarkers for depression with postpartum onsetReference Mehta, Newport, Frishman, Kraus, Rex-Haffner and Ritchie11 and the GnRHa model;Reference Frokjaer, Pinborg, Holst, Overgaard, Henningsson and Heede6 there was no participant overlap between these two studies. This enabled us to determine whether transcriptional and epigenetic sensitivity to sex steroid-hormone manipulation of clinically derived genes map onto depressive symptoms and related SERT brain binding in a controlled pharmacological study. For this we assessed longitudinal gene-expression and DNA-methylation profiles of the 116 identified transcripts to predict PND in women undergoing GnRHa intervention or placebo.
Methods and materials
Participants
Healthy women were recruited via an internet advertisement and invited to a face-to-face screening by trained clinicians. Women with no history of neurological or psychiatric disorders (ICD-10 [1992] or DSM-IV [1994] Axis-I diagnostic criteria for obsessive–compulsive disorder, anxiety, major depression, bipolar disorder/mania or schizophrenia [Schedules for Clinical Assessment in Neuropsychiatry 2.1 interview]), no premenstrual dysphoric disorder (DSM-IV criteria), no alcohol misuse or illegal drug use, and a normal neurological and gynaecological examination were included in the study, as detailed in Frokjaer et al.Reference Frokjaer, Pinborg, Holst, Overgaard, Henningsson and Heede6 Participants scored within Danish norms on the Big Five Personality questionnaire (Revised NEO Personal Inventory) except for the ‘openness’ dimension where they scored higher, as expected for individuals volunteering for a PET study (127 ± 18 versus 107 ± 19, mean ± s.d., P = 0.0001).Reference Frokjaer, Pinborg, Holst, Overgaard, Henningsson and Heede6
A total of 63 healthy women participated in this randomised, placebo-controlled and double-blinded intervention study. Data sets from 60 participants were available for analyses. Of the remaining three participants, one did not receive intervention due to anovulation, one became pregnant and did not complete follow-up and one participant was discontinued in the programme due to technical error with brain imaging data. All participants had regular menstrual cycles and normal hormone status. Baseline assessments were performed in the mid-follicular phase when ovarian hormone levels are most stable and the time since the postovulatory oestradiol drop is maximised. Contingent upon ovulation in their natural cycle as confirmed by transvaginal ultrasound, participants received a subcutaneous injection of a GnRHa implant (ZOLADEX® containing 3.6 mg of goserelin; AstraZeneca, London, UK) (n = 30) or saline (n = 30) in the midluteal phase of their cycle, i.e. on day 22.6 ± 2.5. This timing adhered to the reproductive care guidelines for GnRHa use and allowed a matched timing of menstrual bleeding (placebo group) and withdrawal bleeding (GnRHa group), which enabled blinding. Follow-up (time point 2) was placed post-bleeding at a time point late enough to allow the GnRHa group to have entered their early ovarian suppression phase (16.2 ± 2.6 days after intervention). In a subgroup of the latter-enrolled 38 participants, an extra blood sample was also withdrawn in the initial stimulation phase of GnRHa (follow-up point 1 placed 3–5 days after GnRHa intervention) to capture peripheral features of the stimulatory phase of GnRHa treatment. Brain imaging, clinical rating with the HRSD-17 and collection of DNA was performed at baseline and at time point 2 exclusively (Fig. 1).
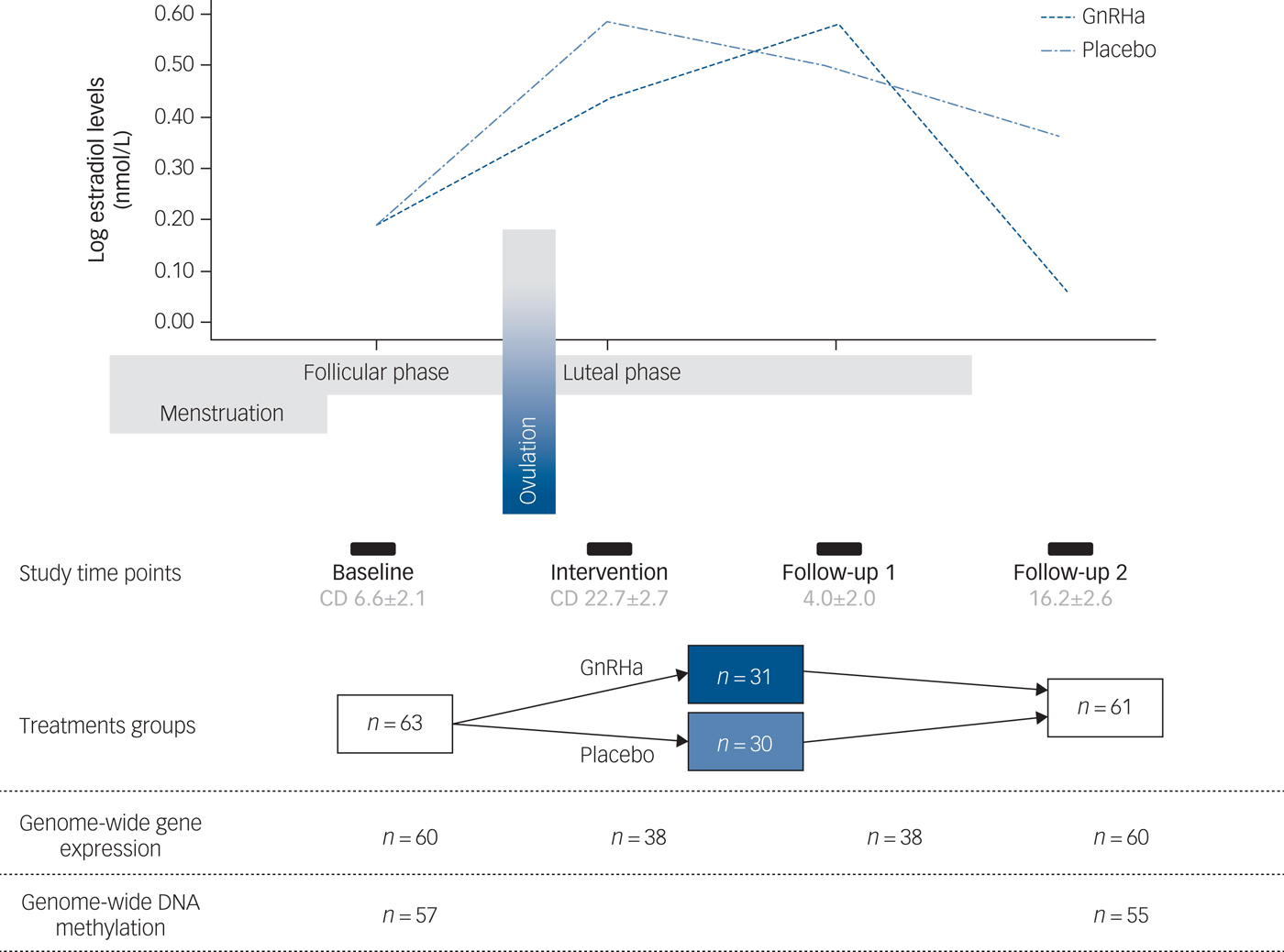
Fig. 1 Study design depicting the different time points and samples available. The follow-up time point 1 depicts the early stimulatory phase of the gonadotrophin-releasing hormone agonist (GnRHa) intervention, whereas time point 2 depicts the subsequent suppression phase. The line plot of the log oestradiol levels in the women is shown across the study at the different time points. The bars indicate time points, i.e. baseline assessment placed in follicular phase at cycle day (CD) 6.6 ± 2.1 (mean ± s.d.), intervention in luteal phase at cycle day 22.7 ± 2.7, early follow-up (follow-up 1) at 4.0 ± 2.0 days post-intervention and late follow-up (follow-up 2) at 16.2 ± 2.6 days post-intervention.
All participants gave written informed consent. The study was approved by the ethics committee (protocol identifier: H-2-2010-108), and registered at www.clinicaltrials.gov under the code NCT02661789.
Experimental procedures
DNA and RNA collection
Samples were stored at −20°C. RNA was isolated from peripheral blood collected in PAXgene tubes using the PAXgene Blood miRNA kit (PreAnalytiX, QIAGEN, Germany). RNA integrity and concentration were analysed using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, US); the RNA integrity number (RIN) ranged from 6.6–9.4, with a mean (s.d.) RIN of 8.4 (0.4). The RNA samples were sent to the Max Planck Institute of Psychiatry to run on microarrays as previously described.Reference Mehta, Newport, Frishman, Kraus, Rex-Haffner and Ritchie11 Illumina's GenomeStudio version 2011.1 with Methylation module 1.9.0 software (Illumina Inc, San Diego, CA, USA, https://support.illumina.com/downloads/genomestudio_software_20111.html), with default Illumina settings, was used for the analysis.
Oestradiol concentrations
Serum oestradiol concentrations were determined as previously described.Reference Frokjaer, Pinborg, Holst, Overgaard, Henningsson and Heede6
SERT imaging
Brain SERT binding was imaged by using [11C]DASB-PET based on 90-minute dynamic acquisition immediately after bolus injection. Further details on [11C]DASB imaging and quantification were previously described.Reference Frokjaer, Pinborg, Holst, Overgaard, Henningsson and Heede6, Reference Frokjaer, Vinberg, Erritzoe, Svarer, Baare and Budtz-Joergensen12
Statistical analyses
Raw data were exported using the Illumina Beadstudio program (Illumina Inc, San Diego, CA, USA, https://support.illumina.com/downloads/genomestudio_software_20111.html) and loaded into R for downstream analysis (http://www.R-project.org). Samples with probe detection call rates less than 95% and those with an average intensity value of either under 50% of the experiment-wide sample mean or less than 2000 arbitrary units were excluded from further analysis.
Gene expression data were transformed and normalised using the variance stabilising normalisation.Reference Huber, von Heydebreck, Sultmann, Poustka and Vingron13 Probes passing the filter criteria of Illumina probe detection P-value of less than 0.01 in at least one sample were used for subsequent analysis. We used the surrogate variable analysis (SVA) method to account for known and unknown factors such as batch effects by including them as covariates in the model, controlling for potential bias. After quality control and filtering, a total of 32 143 gene-expression probes were used for analysis.
For DNA methylation, the data were background and control normalised using the minfi package.Reference Aryee, Jaffe, Corrada-Bravo, Ladd-Acosta, Feinberg and Hansen14 Subset-quantile Within-Array Normalisation was used to remove technical differences between Infinium-I and -II probes via the minfi package. The methylation status for each probe was recorded as a β-value ranging between 0 and 1. Probes where more than 50% of the samples had a detection P-value greater than 0.05, probes on the Y chromosome, and those with single-nucleotide polymorphisms within 50 bp from the query site were removed. This resulted in 482 227 CpG (5′-cysteine–phosphate–guanine-3′) probes that were used for subsequent analysis.
Data were analysed using an established analysis pipeline comprising custom statistical programs and scriptsReference Mehta, Klengel, Conneely, Smith, Altmann and Pace15 written in R and Linux. Surrogate variable analyses revealed seven significant SVA vectors which were used as covariates in the model to correct for technical artifacts and hidden confounds. Linear mixed-effects models were used to investigate differential gene expression and DNA methylation and to test their association with phenotypes as repeated measures, after correcting for the SVAs. Analysis-of-variance models were used to extract the P-values. As described previously,Reference Frokjaer, Pinborg, Holst, Overgaard, Henningsson and Heede6 oestradiol concentrations were log-transformed for analysis. For candidate analysis, we used the nominal P-value. The hypergeometric test was used to test for the enrichment and was performed in R. This test helps assess if the observation is indeed statistically significant, i.e. beyond what is expected by chance.
Results
Demographics
The groups were well matched demographically, i.e. age, body mass index and education, as well as for subclinical psychopathology and follicle-stimulating hormone levels (P > 0.05).Reference Frokjaer, Pinborg, Holst, Overgaard, Henningsson and Heede6 Supplementary Table 1 available at https://doi.org/10.1192/bjp.2018.234 shows the clinical profile. Details on genome-wide gene-expression and DNA-methylation samples available at different time points are depicted in Fig. 1.
Gene expression and DNA methylation of the 116-gene PND panel in the GnRHa model
Post-GnRHa-induced gene-expression and DNA-methylation changes within the 116 PND genes (intervention group × time)
To mirror the results from the third trimester of pregnancy,Reference Mehta, Newport, Frishman, Kraus, Rex-Haffner and Ritchie11 we specifically assessed differences in gene expression between baseline and the early post-GnRHa intervention time point (follow-up 1), which provides a model proxy for a hormone-stimulated phase. For the 116 PND transcripts, we tested if associations between changes in gene-expression profiles across baseline to early follow-up post-GnRHa induction were moderated by the intervention (intervention × time point interaction). We observed that 22 of the 116 transcripts (19%) showed significant changes in gene expression (P < 0.05) between baseline and early post-GnRHa intervention (follow-up 1) across the GnRHa and placebo group, this was significant relative to what would be expected by chance (1.7-fold enrichment, P = 0.02). Of these, 10 transcripts also showed persistent changes in gene expression from baseline until the later post-GnRHa time point (follow-up 2). Among these 10 transcripts there were 6 (see Supplementary Table 4 for details) showing a significant change in this cohort at second follow-up; they were also in the same direction as in the PND cohort by Mehta et al,Reference Mehta, Newport, Frishman, Kraus, Rex-Haffner and Ritchie11 indicating that the gene-expression changes post-GnRHa intervention were persistent and had likely ‘carry-over’ effects from the stimulatory phase into the subsequent suppression or hormonal-withdrawal phase.
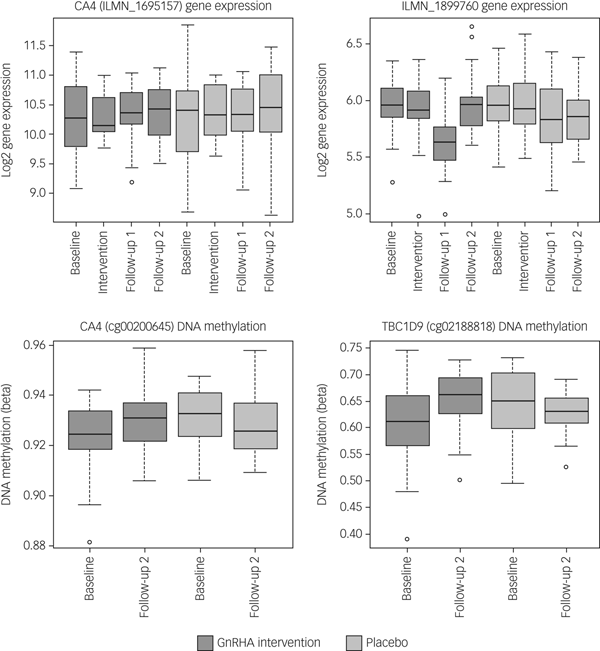
Next, we tested whether the GnRHa treatment also elicited changes in DNA methylation in loci encoding these 116 transcripts. DNA methylation was available at baseline and the later, second follow-up post-GnRHa intervention; hence the DNA-methylation differences represent longer-lasting changes post-GnRHa rather than immediate changes. From the 116 transcripts, 98 had at least one CpG within 1.5 kb of the gene locus on the 450k DNA-methylation array available for analysis after quality control and filtering. For the 2756 CpG sites within these 98 transcripts, analysis was performed to identify those CpGs significant for intervention group × time, using P < 0.05. DNA-methylation levels for the CpG sites within these genes were regressed against the group status (intervention/placebo) × time (baseline to post-GnRHa intervention follow-up 2) (n = 60 women). From those tested, 48 transcripts (49%, 208 CpG sites) had at least one CpG with P < 0.05 (2.6-fold enrichment, P = 1.6 × 10−5; Supplementary Table 5). The difference in DNA methylation from baseline to the later follow-up ranged from −4 to +8%, with 196 (95%) CpGs showing an increase in DNA methylation (average 4% [1%]) and 11 CpGs showing a decrease in methylation (average 3% [1%]). Among the 22 transcripts that had significant gene-expression changes, the majority (n = 18) also had significant changes in DNA methylation (Table 1). Figure 2 depicts examples of gene-expression and DNA-methylation changes.
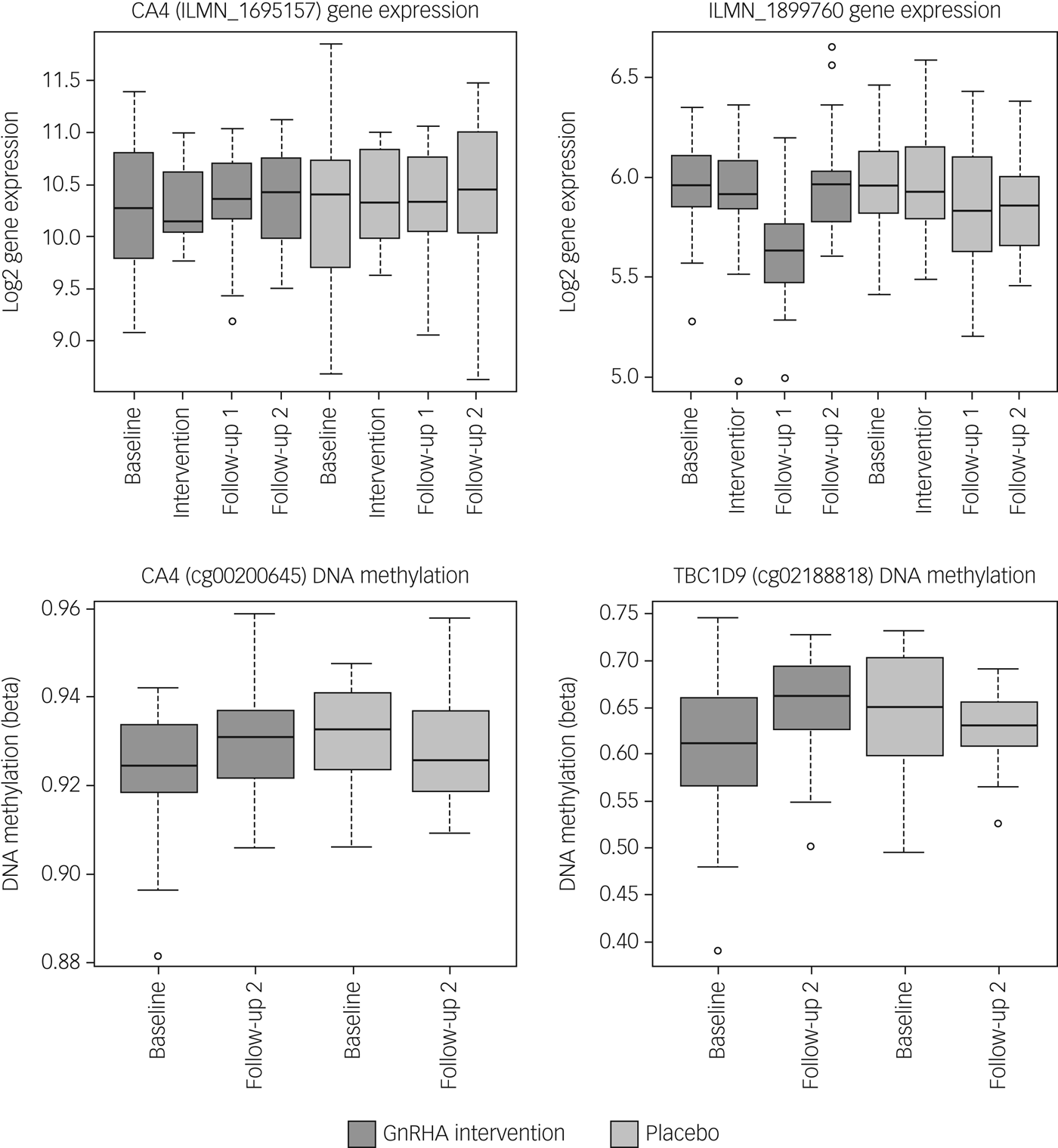
Fig. 2 Gene-expression and DNA-methylation changes post-GnRHa treatment. Top panel indicates gene-expression changes and bottom panel indicates DNA-methylation changes in 2 of the 116 PND genes. Left panel indicates both gene expression for CA4 (P = 0.04) and the DNA methylation (P = 0.023) that were significant. Right panel indicates other examples of an intergenic probe (P = 0.004) that was significant at the level of gene expression (top) and DNA methylation for a CpG within the TBC1D9 gene (bottom) that was significant (P = 0.004). ILMN_1899760 was differentially expressed in the same direction as the previous PND study. CA4 and TBC1D9 showed both significant gene-expression and DNA-methylation changes.
Table 1 A list of the 22 transcripts that had significant gene expression changes
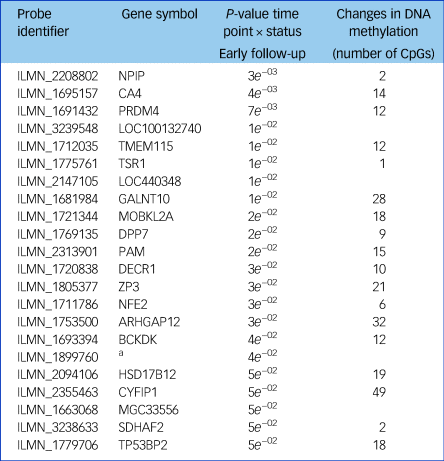
Of these, 18 also had significant changes in DNA methylation after treatment with a gonadotrophin-releasing hormone agonist across the intervention and placebo groups.
a. No gene symbol available.
PND genes associated with oestradiol levels within the GnRHa treatment group
Our previous work demonstrated that GnRHa-triggered depressive symptoms were significantly associated with magnitude of oestradiol decline among the GnRHa-treated women.Reference Frokjaer, Pinborg, Holst, Overgaard, Henningsson and Heede6 To test if the observed changes in gene expression or DNA methylation were driven by fluctuations in oestradiol in response to GnRHa, i.e. proxy for sensitivity to GnRHa, we assessed the association between gene-expression/DNA-methylation changes and changes in oestradiol levels. In terms of gene-expression changes from baseline to early follow-up, 57 of the 116 transcripts (49%) were significantly associated with changes in oestradiol levels at the later follow-up, and 16 transcripts (14%) were associated with early oestrogen changes at the early follow-up (Table 2 and Supplementary Table 3). In terms of DNA-methylation, changes in 76 loci (66%, 368 CpG sites, twofold enrichment, P = 1.5 × 10−4) were significantly associated with the change in oestradiol levels between baseline to late follow-up (Supplementary Table 2).
Table 2 Gene expression changes associated with oestradiol, depressive symptoms and serotonin binding in neocortex within the 116 perinatal-depression transcripts
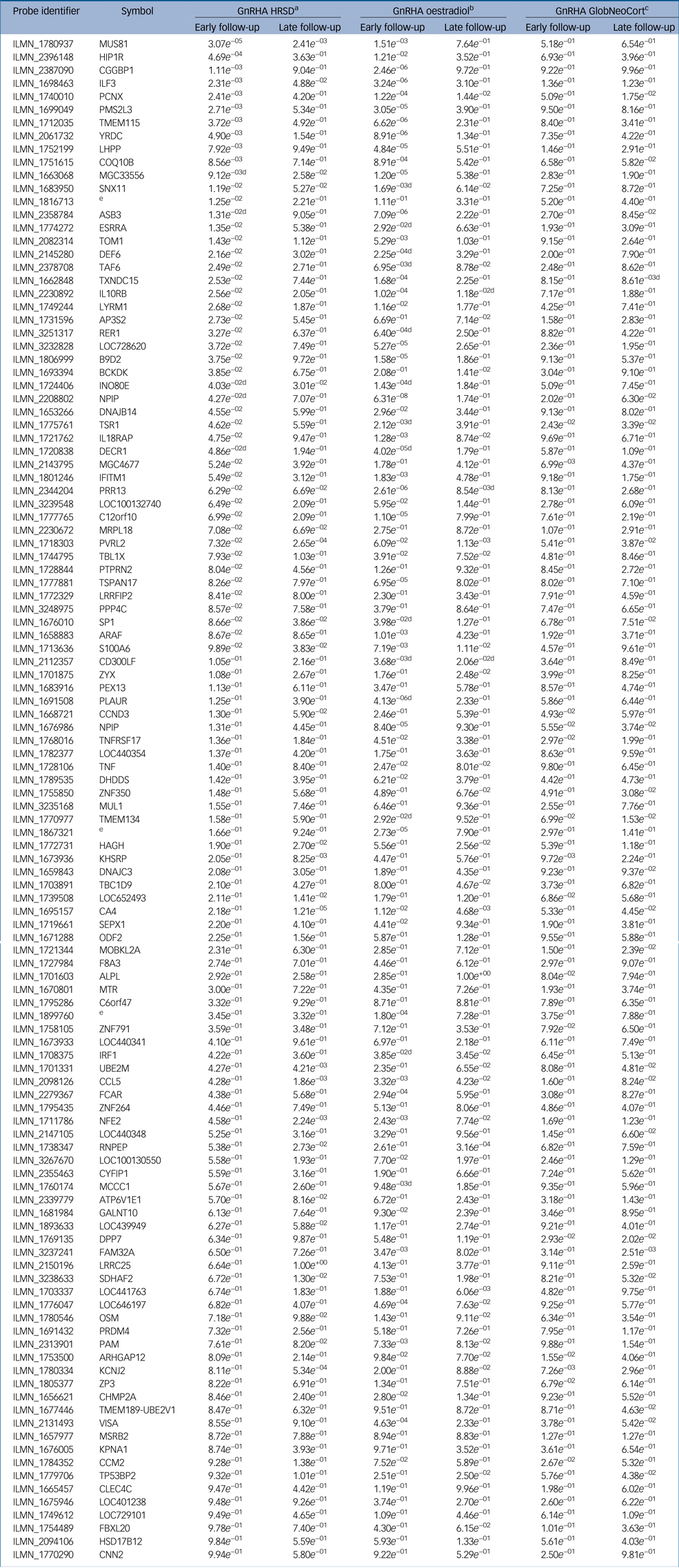
The results shown are from the treatment group. ‘Early’ indicates at early follow-up post-intervention with gonadotrophin-releasing hormone agonist; ‘late’ indicates the late post-intervention follow-up. GnRHA, gonadotrophin-releasing hormone agonist; HRSD, Hamilton Rating Scale for Depression; GlobCortNeo, global neocortex.
a. Indicates genes associated with changes in depressive symptoms.
b. Indicates genes associated with changes in oestradiol symptoms.
c. Indicates genes associated with changes in serotonin binding in the global neocortex.
d. Indicates gene-expression changes that are also associated in the placebo group.
e. No gene symbol available.
PND genes associated with HRSD-17 within the GnRHa treatment group
We had previously demonstrated that GnRHa intervention precipitated depressive symptoms in some women,Reference Frokjaer, Pinborg, Holst, Overgaard, Henningsson and Heede6 hence we tested if gene-expression/DNA-methylation changes between baseline and the two post-GnRHa time points were associated with changes in HRSD-17 scores. For 33 of the 116 transcripts (28%, twofold enrichment, P = 4.4 × 10−7; Table 2) gene-expression changes between baseline and early follow-up were nominally significantly associated with changes in HRSD-17 scores to the early follow-up. For 18 of 116 genes (16%, fold enrichment P > 0.05), early gene-expression changes were significantly associated with mood symptoms at the late follow-up (Table 2). In addition, the DNA-methylation changes from baseline to late follow-up of 77 transcripts (66%, 319 CpG sites) were also associated with changes in HRSD-17 scores from baseline to late post-GnRHa (follow-up 2) (twofold enrichment, P = 2.5 × 10−4; Supplementary Table 2).
PND genes associated with SERT binding in neocortex within the GnRHa treatment group
In this cohort GnRHa induced increases in depressive scores, which were significantly associated with increases in neocortical SERT binding within the GnRHa group only in contrast to placebo.Reference Frokjaer, Pinborg, Holst, Overgaard, Henningsson and Heede6 We therefore tested whether neocortical SERT binding would also associate with gene-expression and DNA-methylation changes in these genes post-GnRHa treatment. We found that of the 116 PND genes, gene expression of 9 transcripts (8%) and 16 transcripts (14%) were associated with changes in SERT binding in neocortex from baseline to early and late post-GnRHa follow-up, respectively (Table 2). Similarly, DNA methylation of 52 transcripts (45%, 156 CpG sites) were also associated with changes in SERT binding in neocortex from baseline to late post-GnRHa (follow-up 2) (twofold enrichment, P = 9.9 × 10−6; Supplementary Table 2).
Discussion
We provide the first direct human evidence of the role of oestrogen fluctuations and their biological consequences as a risk factor for development of hormone-induced depression.
We found (a) that the 116 PND genes were regulated over the time course of the GnRHa intervention on the gene-expression and DNA-methylation level, which correlated with GnRHa-induced changes in oestradiol; and (b) that their dynamic changes predicted changes in HRSD-17 score and SERT brain binding. Our data suggest that dynamic changes in gene expression and DNA methylation in peripheral blood of transcripts, which are regulated over pregnancy and are predictive of PND, also predict mood changes in a pharmacological model. Our results thus directly support a coupling of dynamic sensitivity to oestrogens on the molecular level and sensitivity to develop depressive symptoms across hormonal transitions.
Of the 116 PND-predictive transcripts, we found that a significant proportion of genes were also differentially expressed between baseline and early post-intervention (stimulation phase) across the GnRHa intervention relative to the placebo group. For a proportion of these transcripts, changes were still present at the later follow-up (16 days post-intervention), indicating more long-lasting gene-expression changes carried over from stimulation to the (early) hormonal suppression phase. The genes included NPIP, belonging to a family of a series of primate-specific nuclear pore complex-interacting protein (NPIP) sequences with unknown function but which are well conserved from African apes to humans. Other genes included ZP3, whose protein is a structural component of the zona pellucida and functions in primary binding and induction of the sperm acrosome reaction. Mutations within the ZP3 gene have been reported to be involved in human fertility.Reference Liu, Li, Bai, Yin, Tang and Chi16 Furthermore, we observed that the extent of gene-expression changes in relationship to changes in oestradiol levels also associated with the occurrence of mood symptoms, suggesting that the transcripts identified to predict PND may more generally serve as dynamic biomarker for hormone-induced mood changes, even in a pharmacological model. This raises the question of whether this candidate biomarker can identify women who display oestrogen sensitivity, even outside the pregnancy-stimulated state. Future studies are warranted to test if short-term stimulation, e.g. of blood ex vivo, can unmask the biomarker. Among genes significantly associated with changes in depressive symptoms and oestradiol were several genes related to the immune system such as ILF3, IL10RB and IL18RAP. Another gene was LHPP which encodes an enzyme known as phospholysine phosphohistidine inorganic pyrophosphate phosphatase. Polymorphisms in the LHPP gene together with functional interaction with serotonin receptor 1A (HTR1A) have been previously shown to be associated with MDDReference Knowles, Kent, McKay, Sprooten, Mathias and Curran17 and were associated with resting state brain activity in MDD.Reference Cui, Gong, Tang, Kong, Chang and Geng18 Another interesting gene was the ESRRA gene encoding a nuclear receptor closely related to the oestrogen receptor. ESRRA is expressed in the central nervous system and is required for induction of mitochondrial biogenesis in the brain.Reference Strum, Shehee, Virley, Richardson, Mattie and Selley19 It may regulate multiple metabolic processes critical to neuronal function including mediating expression of brain-derived neurotrophic factor and other target genes, monoamine oxidase A and B, that regulate neurotransmitter metabolism and behaviour.
Oestrogen receptors serve as transcription factors by regulating expression of target genesReference O'Malley8 to mediate the physiological effects of oestrogen. Oestrogen itself can also induce local changes in DNA methylation, which may lead to more lasting regulatory effects of changing sex-steroid levels. We found that changes in DNA methylation within the 116 target genes were even more pronounced than changes in gene expression, with a large proportion of genes differentially methylated from baseline to the post-GnRHa intervention time point. Both increases and decreases in DNA methylation were observed. Changes in DNA methylation over the intervention were associated with both changes in oestrogen levels and depressive symptoms. These results demonstrate that sex-steroid changes induce lasting changes in both gene expression and DNA methylation, and that DNA-methylation changes are more pervasive. Moreover, DNA-methylation changes that occur without concurrent gene-expression changes could be related to poised states of these genes following GnRHa stimulation, reflecting an altered responsivity to oestrogen once stimulated.
Inter-individual differences in dynamic changes of gene expression and DNA methylation that associated with a different susceptibility in hormone-induced mood changes could relate to genetic differences in oestrogen-sensitive enhancers of these target genes. In fact, we have previously shown that genetic variants that alter the transcriptional responsivity to glucocorticoids in peripheral blood associate with an altered amygdala response in a threat task and an increased risk for depression as well as schizophrenia.Reference Arloth, Bogdan, Weber, Frishman, Menke and Wagner20 A different set of genetic variants located in oestrogen-responsive enhancers could be risk factors for oestrogen-induced mood changes. In addition, exposure to adverse life events, a known risk factor for the development of hormone-related mood disorders,Reference Mehta, Quast, Fasching, Seifert, Voigt and Beckmann21 has also been shown to associate with lasting changes in DNA methylation,Reference Klengel, Pape, Binder and Mehta22 which could in turn shape future transcriptional and epigenetic responses.
One of the systems downstream of oestrogen activation is the serotonergic system and – intriguingly – human, rodent and non-human primate studies suggest that oestrogens might influence mood symptoms via effects on this system, as also recently demonstrated in humans.Reference Frokjaer, Pinborg, Holst, Overgaard, Henningsson and Heede6 Further support for this comes from molecular brain imaging studies that indicate a compromised monoamine brain signalling in the depressed state, including in postpartum depression,Reference Sacher, Wilson, Houle, Rusjan, Hassan and Bloomfield23 and from the efficacy of selective serotonin reuptake inhibitors in PND,Reference Molyneaux, Howard, McGeown, Karia and Trevillion24 a main class of antidepressants that selectively increase serotonin in the synapse. In line with our earlier findings,Reference Guintivano, Arad, Gould, Payne and Kaminsky10 we observed significant associations between gene-expression changes of the 116 PND genes (from baseline to early follow-up) with neocortical SERT binding, and several transcripts showed persistent associations with SERT binding even at the later follow-up. Similar, more pronounced, effects were observed for DNA-methylation changes. We speculate that during postpartum, sharply dropping oestrogen levels may further add to serotonergic deficiency and trigger other neurobiological changes, such as disturbed neurogenesis, that may increase risk for depression. Such effects, including oestrogen level-dependent changes in serotonergic signalling, are likely to be more accentuated in women with a heightened sensitivity to oestrogen as discussed previously.Reference Mehta, Newport, Frishman, Kraus, Rex-Haffner and Ritchie11 These findings are interesting in light of a known crosstalk between sex hormones and neurotransmitter systems, thereby providing a potential mechanism for sex hormones to affect brain structures and functions that are central in the pathophysiology of depression.Reference Barth, Villringer and Sacher5
Given that our observations extended from PND to a pharmacological model, our set of biomarkers may also apply to other hormonal transition phases in women's lives, and thus could help characterise an important clinical subgroup of women at risk for hormone-induced depression. Future studies will be necessary to determine if these biomarkers can in fact identify such a subgroup and thus inform personalised prevention or treatment.
This study has several caveats. Although the longitudinal nature of the study is largely an advantage, a few samples had missing data, thereby reducing the power of the study. Replication of these findings in a larger cohort is warranted. Moreover, the gene-expression profiles were measured at baseline, intervention, early and late follow-up, whereas the DNA-methylation profiles were only measured at baseline and later follow-up, thereby restricting the interpretation of the DNA-methylation data and possibility to link changes in DNA methylation with changes in gene expression. Cell-count differences might confound some of our findings, but we have used the SVA method to account for known and unknown factors. Lastly, the GnRHa cohort displayed only subtle depressive symptoms, so one might speculate whether these clinical manifestations are relevant in a diagnostic context or whether they represent correlates of more non-pathological phenomena, such as postpartum blues or unspecific side effects. Nevertheless, the GnRHa-induced symptoms mapped onto the biomarker-associated states of a manifest depression with postpartum onset in the clinical cohort and, therefore, validate GnRHa exposure as a useful means of modelling mechanisms by which hormonal transition can trigger depressive episodes in certain sensitive women. Future studies in clinical populations must elucidate to what extent different diagnostic criteria for depressive episodes and onset specifiers across the peripartum can be discriminated by their gene-expression and epigenetic profiles. As for the function of the specific genes beyond being biomarkers for oestradiol sensitivity, the observed gene-expression signatures of GnRHa were derived from peripheral blood and therefore we interpret their potential direct effects in the central nervous system with caution.
Supplementary material
Supplementary material is available online at https://doi.org/10.1192/bjp.2018.234.
Funding
This study was funded by the Danish Council for Independent research, The Lundbeck Foundation (Center for Integrated Molecular Brain Imaging) and The Capital Region of Denmark, Foundation of Health Research. D.M. is funded by a Vice Chancellors Research Fellowship from Queensland University of Technology.
Acknowledgements
We thank Dorthe Givard, Agnete Dyssegaard, Jacob Madsen, Bente Dall, Sune Keller, Svitlana Olsen and Gerda Thomsen for superb technical assistance. We thank Jessica Keverne for help with proofreading.






eLetters
No eLetters have been published for this article.