1. Synthetic circuits for reprogramming gene expression
The engineering of plants relies heavily on manipulating gene expression to confer advantageous agronomic traits, such as enhanced growth, increased yield or improved tolerance to biotic and abiotic stresses. The direct manipulation of such plant properties has commonly been achieved through the use of well-established tools for reprogramming transcriptional activity and genome output in plants. For instance, over-expressing individual genes in plants has often used regulatory elements such as the 35S promoter from Cauliflower Mosaic Virus (CaMV 35S) (Ow et al., Reference Ow, Jacobs and Howell1987), or the transcriptional activation domain (TAD) from the herpes simplex viral activator VP16 (Wilde et al., Reference Wilde, Cooke, Brammar and Schuch1994). However, strong, ubiquitous and constitutive expression of transgenes provides only rudimentary control of transcription that lacks the nuanced spatiotemporal control of expression that commonly underpins normal biochemical, cellular and developmental properties. This can lead to a variety of deleterious effects in plants, such as metabolic burden and spatiotemporally inappropriate activity, and as a result, the benefit from the desired trait may not be realised due to negative effects on fitness (Cuzick et al., Reference Cuzick, Maguire and Hammond-Kosack2009; Jeong & Jung, Reference Jeong and Jung2015; Kidd et al., Reference Kidd, Kadoo, Dombrecht, Tekeoglu, Gardiner, Thatcher, Aitken, Schenk, Manners and Kazan2011; Nakashima et al., Reference Nakashima, Tran, Van Nguyen, Fujita, Maruyama, Todaka, Ito, Hayashi, Shinozaki and Yamaguchi-Shinozaki2007; Su & Wu, Reference Su and Wu2004; Thatcher et al., Reference Thatcher, Manners and Kazan2009). While a variety of cell-type-specific and inducible promoters (Aoyama & Chua, Reference Aoyama and Chua1997; Zuo et al., Reference Zuo, Niu and Chua2000) have been identified and developed to improve the specificity of expression, these are inherently constrained to their evolved or engineered repertoire of activity patterns. Ideally, expression control systems would have the ability to integrate an arbitrary number of signals to drive any desired spatiotemporal gene expression patterns.
The implementation of synthetic gene circuits in plants has the potential to address these challenges by enabling the construction of customisable logical operations that underpin extensive programmability in gene expression. Over the past two decades, a wide range of gene circuit systems have been developed and implemented in bacteria, yeast and cultured mammalian cells, enabling the introduction of new behaviours and cellular functions (Andres et al., Reference Andres, Blomeier and Zurbriggen2019; Bonnet et al., Reference Bonnet, Yin, Ortiz, Subsoontorn and Endy2013; Brophy & Voigt, Reference Brophy and Voigt2014; Elowitz & Leibler, Reference Elowitz and Leibler2000; Gardner et al., Reference Gardner, Cantor and Collins2000). However, the development of synthetic circuits in plants has lagged behind.
Synthetic gene circuits employ biological parts such as DNA, RNA and proteins to sense and integrate inputs and produce an output representing an expression state, according to the programmed logical operation. A synthetic gene circuit consists of three functional modules: sensors, integrators and actuators (Figure 1a). Sensors detect cellular and environmental signals, which are the inputs to integrators that perform a logical operation to compute a specific output signal, which actuators relay to alter cell function (Jusiak et al., Reference Jusiak, Cleto, Perez-Piñera and Lu2016).

Figure 1. Basic components of a synthetic gene circuit and truth tables. (a) The sensor module consists of a promoter modulated by intrinsic or environmental signals to express the inputs, such as recombinases, sequence-specific DNA-binding proteins or gRNAs for dCas9. The Integrator module is an engineered promoter that contains customised binding sites for integrating the inputs to compute a specific output signal. For recombinase-based circuits, the binding sites are either present upstream and downstream of a transcriptional block that is introduced in the 5’ UTR region or upstream and downstream of the integrator. The actuator module transmits the processed signal to achieve the desired change in cellular function. For circuit optimisation, reporter genes are often used circuit outputs, which can subsequently be replaced with a functional actuator such as a transcription factor (TF) or artificial targeted transcriptional regulator that can reprogramme endogenous pathways. (b) Schematic representation of a NOR gate and a CRISPRi-based circuit, illustrating the assembly of sensor, integrator, and actuator modules into a functional two-input NOR gate. (c) Truth tables of different logic gates, where A and B represent two distinct input signals and Q represents the output. The presence and absence of signals are indicated by green and black boxes, respectively. (d) Schematic representation of an OR gate constructed by layering two NOR gates.
To date, three main technologies have been employed to design synthetic gene circuits for programming gene expression in plants: recombinase systems, DNA-binding proteins and CRISPR interference (CRISPRi) (Lloyd et al., Reference Lloyd, Ly, Gong, Pflueger, Swain, Pflueger, Fourie, Khan, Kidd and Lister2022; Brophy et al., Reference Brophy, Magallon, Duan, Zhong, Ramachandran, Kniazev and Dinneny2022; Khan et al., Reference Khan, Herring, Zhu, Oliva, Fourie, Johnston, Zhang, Potter, Pineda, Pflueger, Swain, Pflueger, Lloyd, Secco, Small, Kidd and Lister2024). In a simple gene circuit, the sensor module drives the expression of input components (recombinases, DNA-binding proteins or sgRNAs in CRISPR-dCas9 systems), forming the basis of the circuit’s logic. The integrator module consists of engineered promoter sequences with customisable binding sites for these input-derived components, which interact with the integrator to modulate transcription (Figure 1a). Finally, the actuator module transmits the processed signal from the integrator, altering cellular functions. Reporter genes are typically used as the circuit output during optimisation to evaluate performance. Once optimised, the reporter gene can be replaced by effectors, such as transcription factors (TFs), to regulate the expression of specific endogenous genes (Figure 1a).
These modules can be combined in various configurations to create two-input gates and implement Boolean logic operations, such as a NOR gate. For example, a CRISPRi-based NOR gate can be constructed using two distinct sgRNAs as inputs (Figure 1b). In this example, one sgRNA is driven by a cell-type-specific promoter, while the other is driven by an inducible promoter, and both are targeted to an integrator module containing unique binding sites for these sgRNAs. When either or both sgRNAs are present, dCas9 binds to the integrator, repressing the expression of the output gene. Consequently, the output is only produced in the absence of both input sgRNAs, thereby creating a NOR gate. This configuration allows precise regulation of gene expression in response to environmental signals and specific cellular contexts, demonstrating how synthetic gene circuits can be designed to perform complex logical functions in plants.
One of the major goals of synthetic biology research is to create synthetic gene circuits that function analogously to the logical operations that underpin the fundamental functions of computers and other electronic devices (Miyamoto et al., Reference Miyamoto, Razavi, DeRose and Inoue2013). Similar to endogenous cellular regulatory pathways that sense and integrate multiple input signals to produce specific outputs only under certain conditions, synthetic gene circuits enable the construction of transcriptional programmes that respond to multiple customisable inputs (Medford & Prasad, Reference Medford and Prasad2016). These circuits are based on logic gates, which make use of Boolean algebra operations to integrate multiple input signals into truth values of 1 (true) and 0 (false) that can be linked together to produce distinct logic such as AND, OR, NOT, A NIMPLY B and XOR operations, amongst others (Figure 1c). By constructing logic gates from DNA, RNA, and protein components, these engineering principles can be achieved in living organisms, where specific transcriptional outputs in the form of selective activation or repression of desired transgenes or pathways can be achieved in response to input signals (Weinberg et al., Reference Weinberg, Pham, Caraballo, Lozanoski, Engel, Bhatia and Wong2017). For example, in a simple implementation, an AND gate can be used to achieve highly specific gene expression patterns only when both inputs are simultaneously present. The ability to design and implement complex gene circuits in plants would enable unprecedented precision and customisability in the control of patterns of gene expression, with myriad applications from the dissection of fundamental plant cells and developmental processes to the introduction of novel traits for improved agricultural productivity.
2. Current state of plant synthetic gene circuits
Synthetic biological circuits can be constructed to operate at different molecular levels within a cell, incorporating transcriptional, post-transcriptional, and translational processes. However, to date, the advanced circuit platforms developed in plants have focused on modifying transcriptional levels. In this section, we review the current literature on the construction of advanced plant synthetic circuits.
Based on the technology used for the construction of gene circuits in plants, we can categorise them into two main types: irreversible (i.e. memory) circuits and reversible circuits. Irreversible circuits are designed to retain a memory of an input signal, subsequently maintaining the output state once triggered by the input. In contrast, reversible circuits allow for dynamic changes in the output state in response to varying input signals.
2.1. Memory circuits
Recombinases are enzymes that ‘flip’ or recombine DNA segments between their cognate binding sites, making them ideal for building memory devices since the genetic reconfiguration at the DNA level is maintained permanently without the requirement for the continuous presence of input signals (Bonnet et al., Reference Bonnet, Subsoontorn and Endy2012; Bowyer et al., Reference Bowyer, Zhao, Subsoontorn, Wong, Rosser and Bates2016; Friedland et al., Reference Friedland, Lu, Wang, Shi, Church and Collins2009; Lapique & Benenson, Reference Lapique and Benenson2014; Roquet et al., Reference Roquet, Soleimany, Ferris, Aaronson and Lu2016; Siuti et al., Reference Siuti, Yazbek and Lu2013).
Bernabé-Orts and colleagues developed a memory switch in Nicotiana benthamiana using the bacteriophage PhiC31 serine integrase. By using its cognate recombination directionality factor, the authors were able to reverse the implementation of this memory switch by ~60% when tested in stable transgenic plants (Bernabé-Orts et al., Reference Bernabé-Orts, Quijano-Rubio, Vazquez-Vilar, Mancheño-Bonillo, Moles-Casas, Selma, Gianoglio, Granell and Orzaez2020). Using an expanded repertoire of DNA recombinases, Weinberg and colleagues reported the Boolean Logic and Arithmetic through DNA Excision (BLADE) platform, characterising 12 different recombinases for the construction of 16 Boolean logic gates and complex circuits in human cells (Weinberg et al., Reference Weinberg, Pham, Caraballo, Lozanoski, Engel, Bhatia and Wong2017). Building upon this, we recently developed and optimised a DNA recombinase-based memory gene circuit platform in the model plant Arabidopsis (Lloyd et al., Reference Lloyd, Ly, Gong, Pflueger, Swain, Pflueger, Fourie, Khan, Kidd and Lister2022). Using the Flp and B3 recombinases as two different inputs, we successfully engineered different logic gates within plant cells, including NOT, OR, NOR, AND, NAND and A NIMPLY B gates. To demonstrate the functionality of this system in stable transgenic plants, we created an AND gate in the roots of Arabidopsis, using DEX- and cell-type-specific promoters to drive the expression of Flp and B3 recombinases, respectively, to achieve chemically inducible cell-type-specific control of reporter expression in roots.
Recently, Guiziou and colleagues further expanded the use of serine integrases to record developmental events in Arabidopsis roots (Guiziou et al., Reference Guiziou, Maranas, Chu and Nemhauser2023). The authors used promoters specific to lateral root development to express PhiC31 and Bxb1 serine integrases, allowing for switching between two different fluorescent reporter genes. This integrase-based circuit resulted in the expression of reporter genes during root development, permanently marking all daughter cells. Further exploration of the use of serine integrases in constructing complex memory devices could enable sophisticated new tools for recording diverse plant developmental processes and responses to environmental cues.
These studies demonstrate the utility of DNA recombinases for engineering complex synthetic circuits with customisable logic functions and memory capabilities in plants. However, while recombinases achieve robust control of circuit activity through DNA-level changes, this comes with the inherent limitation in programmable circuits because their logic is irreversible (in the absence of an additional recombination directionality factor). This generally prevents reversible dynamic responsiveness of recombinase-based circuits to variable input stimuli.
2.2. Reversible circuits
Reversible circuits have the ability to dynamically switch between ‘ON’ and ‘OFF’ states based on the input states, enabling transient responses to changing signals.
2.2.1. DNA-binding proteins
DNA-binding proteins that can bind to a specific DNA sequence are ideal components for creating synthetic gene circuits. These proteins can be used as synthetic transcriptional regulators by fusing them to transcriptional activation or repression domains, creating fusion proteins that can be used for targeting natural or modified promoters containing the DNA-binding sequence, causing a change in transcription (Lienert et al., Reference Lienert, Lohmueller, Garg and Silver2014; Weber & Fussenegger, Reference Weber and Fussenegger2009). The use of recently characterised plant transcriptional regulatory domains with DNA-binding proteins will further expand the toolkit of synthetic transcriptional regulators for building gene circuits in plants (Hummel et al., Reference Hummel, Markel, Stefani, Staller and Shih2024; Morffy et al., Reference Morffy, Van den Broeck, Miller, Emenecker, Bryant, Lee, Sageman-Furnas, Wilkinson, Pathak, Kotha, Lam, Mahatma, Pande, Waoo, Wright, Holehouse, Staller, Sozzani and Strader2024).
In a recent study, Brophy and colleagues fused bacterial sequence-specific DNA-binding proteins to activator and repressor domains, which were coupled to engineered promoters, to develop a comprehensive set of logic gates in plants (Brophy et al., Reference Brophy, Magallon, Duan, Zhong, Ramachandran, Kniazev and Dinneny2022). To evaluate the performance of these logic gates in stable Arabidopsis plants, the inputs (activators or repressors) were expressed under the control of root-specific promoters to drive GFP expression in a cell-type-specific manner. However, transferring these circuits from the leaves of N. benthamiana to the roots of Arabidopsis required further optimisations, suggesting the need for species-specific fine-tuning of biological parts for constructing gene circuits.
DNA-binding proteins, such as zinc finger proteins (ZNFs) and transcription activator-like effectors (TALEs) can have high binding specificity for their target DNA sequences and therefore are useful for creating synthetic TFs (Gaj et al., Reference Gaj, Gersbach and Barbas2013). However, creating TFs based on ZNFs or TALEs can be technically challenging and have higher error rates of construction (Beumer et al., Reference Beumer, Trautman, Christian, Dahlem, Lake, Hawley, Grunwald, Voytas and Carroll2013; Ramirez et al., Reference Ramirez, Foley, Wright, Müller-Lerch, Rahman, Cornu, Winfrey, Sander, Fu, Townsend, Cathomen, Voytas and Joung2008). Distinct ZNFs and TALEs need to be engineered for each different target DNA sequence, and therefore offer less flexibility in terms of programmability to construct complex circuits (Gao et al., Reference Gao, Tsang, Gaba, Wu, Lu and Liu2014). For example, Schreiber and colleagues created an AND gate in N. benthamiana using a split-TALE system, where the TALE DNA-binding domain and the transcription activation domain are separately fused to interacting protein domains (Schreiber et al., Reference Schreiber, Prange, Hoppe and Tissier2019). The AND gate is switched on when the split components of the TALE activation system are reconstituted by the physical interaction of the protein domains. The split-TALE approach is a promising system for protein–protein interaction-based AND gates in planta that may exhibit higher target specificity due to the required simultaneity of DNA binding. However, such split approaches offer less flexibility in designing modular complex gene circuits due to the need for multiple distinct pairs of interacting proteins to create other logical operations.
Bacterial allosteric TFs (aTFs) are regulatory proteins that bind to specific DNA sequences and can be used as transcriptional regulators. Their activity can be modulated by the presence of a ligand or metabolite (Li et al., Reference Li, Li, Tan, Xin and Wang2023). In a recent study, Ferreira and Antunes (Ferreira & Antunes, Reference Ferreira and Antunes2024) utilised bacterial aTFs, combined with engineered promoters, to construct logic gates in plants using transient gene expression assays. Moreover, the authors used phenylpropanoid-related metabolites as inputs, demonstrating that these aTF-based gene circuits can function as biosensors to regulate plant metabolic pathways.
2.2.2. CRISPR-dCas9 System
Nuclease dead versions of the Cas9 protein (dCas9) do not cut the DNA backbone and therefore are able to function as highly customisable DNA-binding proteins for which the target site is determined by the gRNA sequence, providing a highly reprogrammable targeted binding system (Bikard et al., Reference Bikard, Jiang, Samai, Hochschild, Zhang and Marraffini2013; Qi et al., Reference Qi, Larson, Gilbert, Doudna, Weissman, Arkin and Lim2013). dCas9 has been used as a transcriptional activator (CRISPRa) or repressor (CRISPRi) by fusing it to activation or repressor domains, respectively, for regulating gene expression in plants (Lowder et al., Reference Lowder, Paul and Qi2017; Piatek et al., Reference Piatek, Ali, Baazim, Li, Abulfaraj, Al-Shareef, Aouida and Mahfouz2015; Vazquez-Vilar et al., Reference Vazquez-Vilar, Selma and Orzaez2023). With CRISPR-Cas9 or related systems, expression of either the dCas9 component or the targeting gRNA can act as input signals to create synthetic gene circuits. Kar and colleagues demonstrated the construction of CRISPRa-based YES gates in plants. To enable activation of a reporter gene, the authors engineered CaMV 35S-based minimal promoters that could be bound by a dCas9-VP64 activator (Kar et al., Reference Kar, Bordiya, Rodriguez, Kim, Gardner, Gollihar, Sung and Ellington2022). The performance of these YES gates was tested in the leaves of N. benthamiana and Arabidopsis, with the input sgRNAs expressed from CaMV35S and ethylene-inducible promoters, respectively.
Further augmentation of dCas9 can confer additional regulatory capabilities and context specificity. Khakhar and colleagues created hormone-activated Cas9-based repressors (HACRs) by fusing dCas9 to plant hormone-induced degrons and the N-terminal 100 amino acids of the TOPLESS repressor (Khakhar et al., Reference Khakhar, Leydon, Lemmex, Klavins and Nemhauser2018; Szemenyei et al., Reference Szemenyei, Hannon and Long2008). The degrons target the dCas9 repressor for degradation in the presence of their respective plant hormones, conferring sensitivity of the synthetic HACRs to auxin, gibberellins and jasmonates. The auxin-degradable repressor was successfully used to reprogram Arabidopsis auxin signalling by targeting the PIN-FORMED 1(PIN1) gene, which encodes an auxin efflux carrier responsible for root and shoot development. This resulted in production of fewer side branches compared to wild-type plants, an agriculturally relevant trait for higher density planting.
The extreme reconfigurability of dCas9 target DNA binding specificity provides valuable opportunities for creating complex gene circuits. Recently, we reported the construction of the first CRISPRi-based plant gene circuit platform able to compute a range of Boolean logic functions (Khan et al., Reference Khan, Herring, Zhu, Oliva, Fourie, Johnston, Zhang, Potter, Pineda, Pflueger, Swain, Pflueger, Lloyd, Secco, Small, Kidd and Lister2024). A library of engineered TCTP and CaMV 35S promoters was generated to contain two distinct gRNA target sites located immediately upstream and downstream of the promoters’ TATA box. Binding of dCas9 to either or both of the gRNA target sites within these integrators caused strong repression of the promoter activity by CRISPRi, thereby functioning as a NOR gate. Importantly, every Boolean logical operation can be created by linking multiple NOR gates together in different configurations, making it a ‘universal’ gate. By optimising the expression and maturation of input gRNAs from RNA polymerase II promoters, gRNAs could be used as the input signals into the circuit, as well as outputs of integrators, allowing multiple NOR gates to be linked to create multi-layered circuits and more complex Boolean logic functions. For example, an OR gate can be constructed by layering two NOR gates (Figure 1d). The output of the first NOR gate (NOR-1), here denoted sgRNA-C, serves as an input for the second NOR gate (NOR-2). The output of NOR-2 may be either a reporter gene, a TF, or other desired transcriptional outputs. In the absence of input sgRNAs for NOR-1, the output sgRNA-C from NOR-1 is expressed, leading to repression of the NOR-2 output. Conversely, in the presence of input sgRNAs for NOR-1, the output sgRNA-C is not produced due to CRISPRi, allowing the expression of the NOR-2 output. Furthermore, reversible environmentally controlled circuit activity could be demonstrated in stable transgenic Arabidopsis plants with these circuits.
As the DNA targeting specificity of dCas9 can be very easily altered by changing the sgRNA sequences, CRISPR-based circuits exhibit a very high level of programmability, offering some benefits compared to other technologies for creating synthetic circuits. The use of different RNA-guided DNA-binding CRISPR systems such as dCpf1, or alternative technologies including the SunTag (Tanenbaum et al., Reference Tanenbaum, Gilbert, Qi, Weissman and Vale2014), MS2 stem-loops (Konermann et al., Reference Konermann, Brigham, Trevino, Joung, Abudayyeh, Barcena, Hsu, Habib, Gootenberg, Nishimasu, Nureki and Zhang2015) and Casilio (Cheng et al., Reference Cheng, Jillette, Lee, Plaskon, Fujiwara, Wang, Taghbalout and Wang2016) systems with dCas9, could also be implemented to further improve the performance of existing CRISPR-dCas9-based circuits.
2.2.3. RNA-based circuits
In contrast to DNA-based circuits, RNA-based circuits operate at the post-transcriptional level. To date, the development of riboswitches in plants has been limited to single-use designs lacking the ability to be stacked or expanded upon for more complex circuits. The main challenge lies in identifying highly specific RNA-binding proteins or miRNAs, which hinders the construction of sophisticated RNA-based circuits (Matsuura et al., Reference Matsuura, Ono, Kawasaki, Kuang, Fujita and Saito2018).
Liang and colleagues used the CRISPR associated Csy4 system to construct NOT gates in N. benthamiana leaves and rice protoplasts, achieving >400-fold repression of the targeted transgenes (Liang et al., Reference Liang, Richardson, Yan, Benites, Cheng-Yue, Tran, Mortimer, Mukhopadhyay, Keasling, Scheller and Loqué2017). Additionally, the authors demonstrated cell-type-specific and inducible repression with Csy4-based NOT gates in Arabidopsis, and identified additional orthologs of the Csy4 gene (ND02 and MZ1T) that could allow further expansion of RNA circuit capabilities. Though this system results in efficient repression of the target transcripts, Csy4 requires a specific sequence in the mRNA transcript to induce repression, and this sequence cannot be changed, thus limiting its use to fine-tuning of transgenes.
However, recent advances in RNA-targeting CRISPR proteins offer promising prospects to overcome these limitations and enable the development of more advanced RNA-level circuits. RNA-guided RNA-targeting CRISPR systems have emerged as effective tools for gene repression in mammalian systems. Unlike Csy4, some of these CRISPR proteins, for example, CasRx (Konermann et al., Reference Konermann, Lotfy, Brideau, Oki, Shokhirev and Hsu2018), do not rely on any specific target-binding sequences, allowing them to be targeted to any mRNA transcript by provision of a corresponding gRNA (Figure 2a). This characteristic makes them ideal for creating programmable RNA-based complex circuits in plants, as they can be readily programmed by modifying the input sgRNAs (Figure 2a). Furthermore, synthetic circuits based on these CRISPR systems can be easily interfaced with the host cell to target endogenous transcripts.
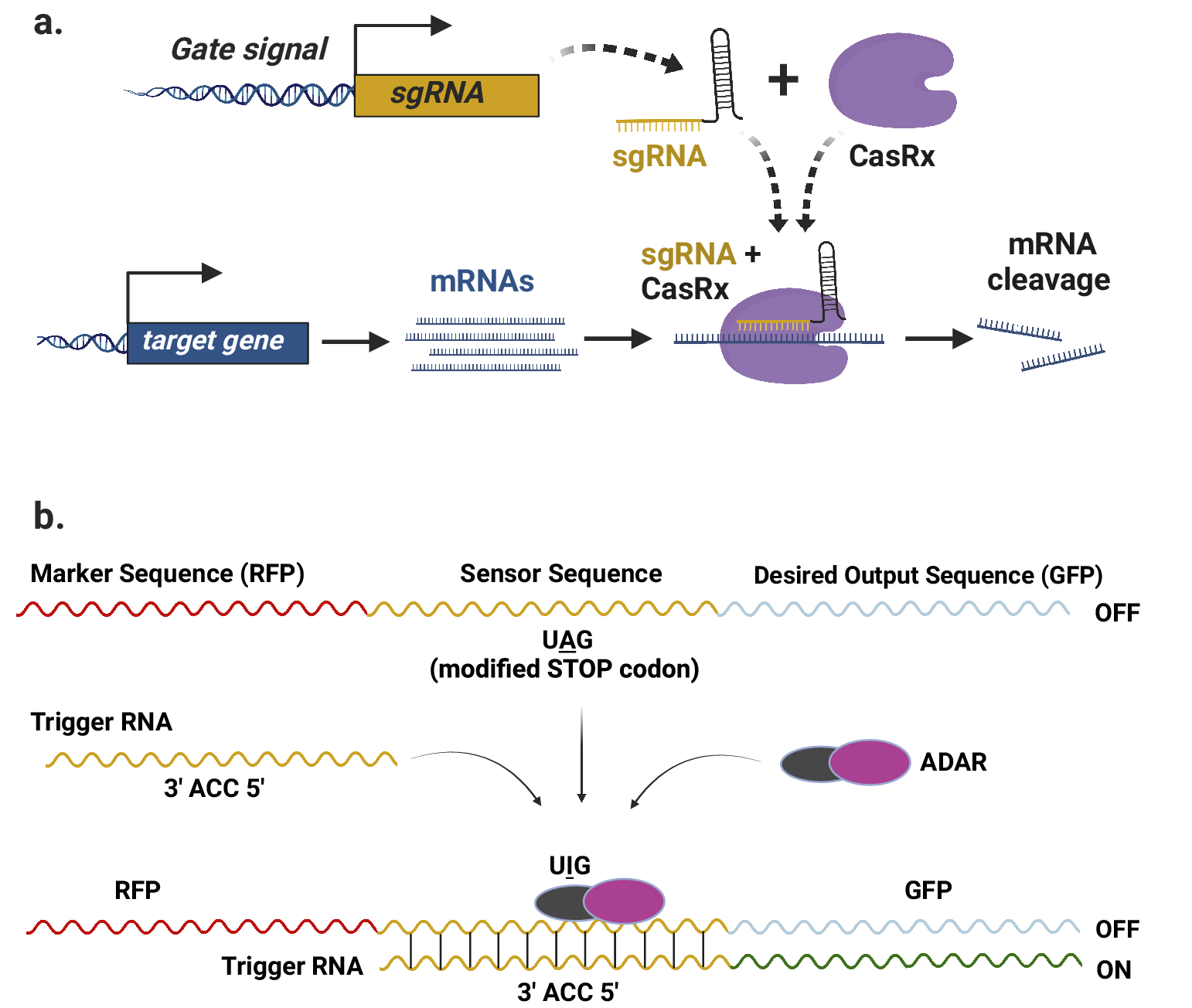
Figure 2. RNA-based circuits in plants. (a) Schematic representation of sgRNA-directed targeted degradation of the mRNA transcripts of a target gene by RNA-guided RNA-targeting CRISPR-CasRx system. The sgRNA can be designed to target any region of the mRNA transcript, as CasRx does not require a PAM sequence. (b) Illustration of the RADAR technology for programmable expression of a desired output such as GFP. An optional sequence, such as RFP, can be included as a marker upstream of the sensor sequence (adapted from (Kaseniit et al., Reference Kaseniit, Katz, Kolber, Call, Wengier, Cody, Sattely and Gao2022).
RNA sensing using adenosine deaminases acting on RNA (RADAR) is a recently reported technology that is programmable, modular and capable of creating cell-type-specific circuits (Kaseniit et al., Reference Kaseniit, Katz, Kolber, Call, Wengier, Cody, Sattely and Gao2022). This approach involves two major components incorporated into a single transcript. First, a sensor sequence that is complementary to an endogenous RNA sequence, known as the trigger sequence, specific to a particular cell type. A ‘UGG’ codon in the sensor sequence is changed to a ‘UAG’ stop codon, halting translation of the downstream output sequence. Second, an output sequence, which is the desired sequence that needs to be expressed in the targeted cell type. As the sequences of sensor and trigger are complementary, a double stranded RNA is formed that is recognised by adenosine deaminase, resulting in the editing of adenosine (A) to Inosine (I) to edit away the UAG stop codon. Thus, translation of the downstream output sequence is enabled (Figure 2b). This technology has been successfully tested in plants, suggesting its potential use in RNA-based circuits for programmable control of translation in specific cell types.
3. Limitations in designing effective circuits
The core engineering principle of synthetic biology is based on the Design-Build-Test-Learn (DBTL) model, which serves as a framework for developing synthetic circuits and gene networks. In the Design phase, genetic components such as genes and regulatory elements are selected and arranged to achieve a specific biological function. Next, in the Build phase, the designed circuits are physically constructed by assembling DNA sequences and introducing them into host organisms. The Test phase involves evaluating the performance of the circuits, measuring outputs, gene expression or protein levels, to gather data. Finally, in the Learn phase, insights are derived from the data to assess whether the circuit functions as expected and to identify areas for improvement, guiding the next round of design refinement. While the DBTL model is a powerful approach to developing synthetic circuits, there are certain limitations that underscore the need for better predictive models and more efficient experimental processes to enhance its effectiveness and scalability (Figure 3). In this section we discuss the key challenges of evaluating circuit performance, high-throughput testing, modelling circuit activity and implementation in crops.
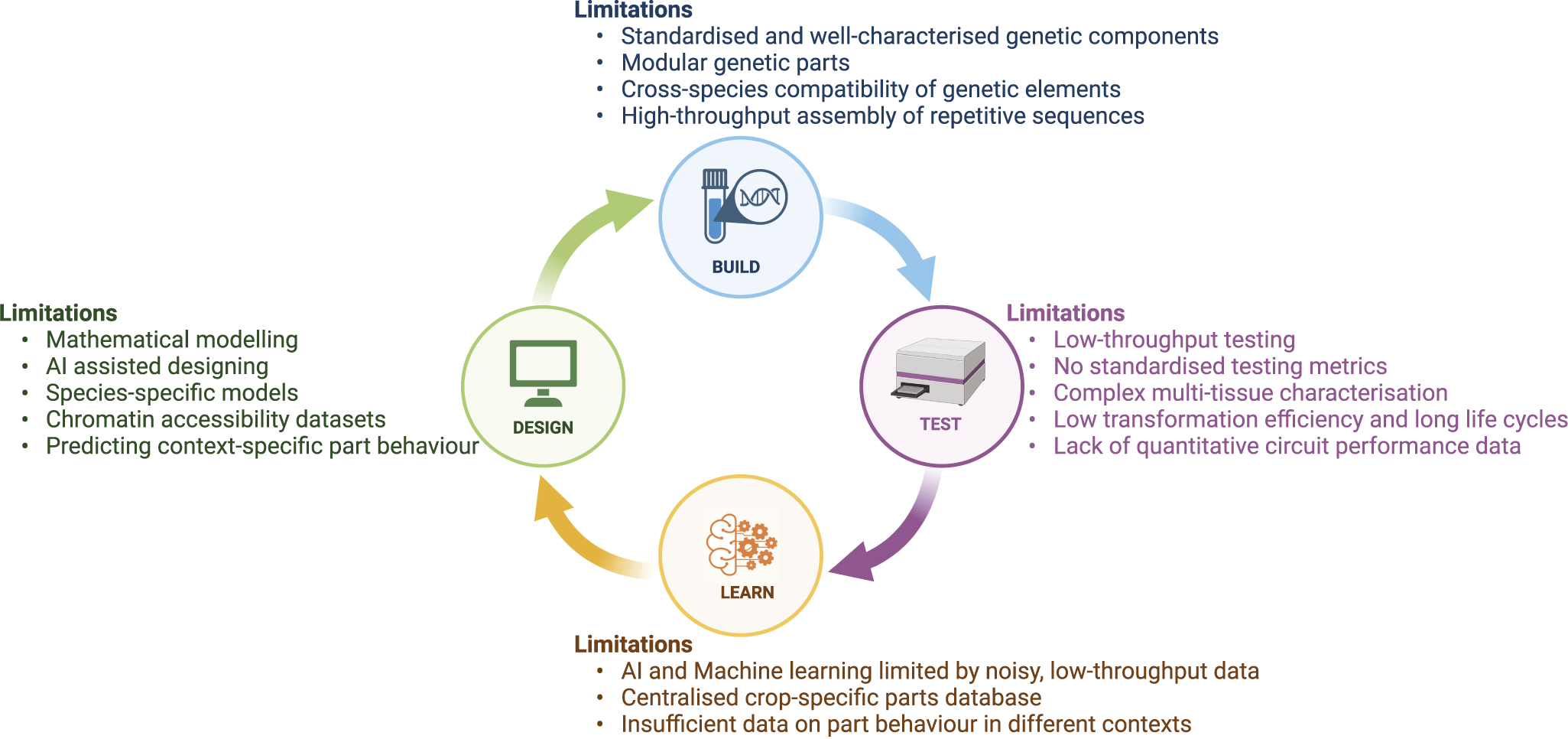
Figure 3. Limitations of the Design-Build-Test-Learn model for implementing synthetic gene circuits in crops. The figure highlights major challenges in each phase of the DBTL cycle, including limited models and datasets in Design, lack of standardised parts in Build, low-throughput testing in Test and noisy data with limited databases in Learn. Addressing these challenges is crucial for enabling synthetic gene circuit applications in crops.
3.1. Quantitative assessment of circuit performance
A major limitation in evaluating the performance of plant synthetic circuits is the frequent use of reporter genes as a proxy for circuit activity, and the lack of high resolution quantitative data on circuit componentry. This limited readout obfuscates the myriad molecular processes that underlie gene circuit functionality, from component transcription and translation, to protein-DNA interactions, to transcriptional regulation. While using fluorescent proteins or luminescent reporter systems provide a convenient and low-cost method of assessing circuit output when screening multiple circuit designs, they provide only indirect measures, often capturing only the final circuit output rather than individual sub-components. Notably, recent work by Csibra and Stan (Csibra & Stan, Reference Csibra and Stan2022) on converting arbitrary fluorescence units into absolute units highlights a promising approach for obtaining more quantitative data, although it has not yet been applied in plants. The absence of quantitative molecular data regarding underlying circuit processes makes it challenging to troubleshoot suboptimal and unpredictable circuit designs. Overcoming this paucity of quantitative circuit activity measurements is a key limitation in their development, use, and advancement.
To improve the optimisation, construction and assessment of successful logic gates, it will be crucial to implement experimental approaches for the accurate quantitation of circuit inputs, intermediary functional components and outputs. This would ideally be conducted frequently over time to examine the temporal features of circuits. In the example of CRISPRi circuits, measuring the transcript abundance of the input gRNAs, circuit output (e.g. reporter) and intermediary gRNAs that connect multi-layered circuits would provide far greater insights into how the quantity of key circuit components relate to its performance or problems, and ultimately aid in determining the design rules for building effective logic gates. Without assessing the levels of such circuit components, it becomes very challenging to predict whether poor circuit performance stems from insufficient production of input signals (e.g. due to a weak sensor) and other circuit components, or inadequate recombination, repression, or activation activities. A recent study measured the level of repressor proteins in synthetic circuits in E. coli using RNA-seq (Gorochowski et al., Reference Gorochowski, Espah Borujeni, Park, Nielsen, Zhang, Der, Gordon and Voigt2017); however, to date, the advanced circuit studies in plants have not implemented a strategy to measure the quantity of inputs, intermediary components or outputs of complex circuits. Furthermore, the ability to measure circuit components at single cell resolution would allow fine-scaled dissection of circuit activity when implemented in stable transgenic plants. Regular generation of such data will be critical for improving the ability to accurately model circuit activity and inform new strategies for improvement.
3.2. High-throughput testing
Testing synthetic gene circuits in plants is made more challenging because of their long life cycles and difficulties in reproducible stable genetic modification. To date, the best available options for less time-intensive testing of gene circuits in plants are transient transformation of protoplasts and agrobacterium-mediated infiltrations. Though protoplast transfection provides a moderate throughput approach for testing circuit performance in a short period of time, the copy number of plasmid DNA delivered to cells is uncontrollable, and will not necessarily reflect the regulatory activities of single copy constructs stably introduced into a chromatinised nuclear genome. Consequently, circuit activity measured in protoplasts could exhibit significant differences in comparison to stable transgenic plant lines.
Agrobacterium-mediated infiltrations offer another widely used approach for transient gene expression in plants, with N. benthamiana being the most common host. While this approach may provide certain advantages over protoplast transfection, such as preserving some tissue context, there is no experimental evidence to suggest that it accurately replicates the regulatory environment of stable plant lines. Further exploration of how well transient systems reflect chromatinised, stable integration environments could enhance our understanding of circuit performance in planta. It is worth noting that, as observed in yeast and mammalian systems, transiently delivered linear or plasmid DNA can associate with nucleosomes, which may influence transcriptional activity (Deniz et al., Reference Deniz, Flores, Battistini, Pérez, Soler-López and Orozco2011; Mladenova et al., Reference Mladenova, Mladenov and Russev2009). Similar to protoplast transfection, the copy number of transferred T-DNA molecules is difficult to control, complicating the quantitative assessment of circuit dynamics. Furthermore, this method is limited by its low-throughput nature and applicability to specific tissues, making it unsuitable for evaluating circuit activity across diverse cell types, which will often be a key consideration of in planta circuit functionality.
The lack of high-throughput and robust testing systems to determine the performance of genetic parts and circuits in plants makes it challenging to test a large number of conditions and circuit design permutations, which is required for developing more sophisticated and robust circuits. The use of robotics and other plant species such as Pyscometrilla patens and Marchantia polymorpha may aid development of higher throughput testing systems for screening gene circuits in vivo due to their short life cycles, ease of growth and reproducible stable genetic modification (Frangedakis et al., Reference Frangedakis, Guzman-Chavez, Rebmann, Markel, Yu, Perraki, Tse, Liu, Rever, Sauret-Gueto, Goffinet, Schneider and Haseloff2021; Rensing et al., Reference Rensing, Goffinet, Meyberg, Wu and Bezanilla2020).
3.3. Modelling of circuits
Mathematical modelling is an important tool for designing synthetic gene circuits, offering insight into their dynamics and stability. Such models are often based on differential equations, stochastic simulations and network analysis, and simulate how various factors impact circuit behaviour (McCallum & Potvin-Trottier, Reference McCallum and Potvin-Trottier2021). Gander and colleagues (Gander et al., Reference Gander, Vrana, Voje, Carothers and Klavins2017) used mathematical modelling to predict the output of interconnected CRISPRi-based NOR gates in yeast, demonstrating that minimal transcriptional leakage could be achieved. Similarly, Santos-Moreno and colleagues (Santos-Moreno et al., Reference Santos-Moreno, Tasiudi, Stelling and Schaerli2020) applied modelling to predict dynamic behaviours, such as oscillations and bistable states, in CRISPRi-based circuits in E. coli. Their model incorporated key parameters like gene expression rates, binding affinities, and unspecific binding to predict circuit behaviour over time. In both studies, experimental validation confirmed the effectiveness of these models in guiding the design of reliable logic circuits.
While synthetic circuits in microbes have made significant progress, the development of plant circuits has been slower, mainly dependent on traditional DBTL cycles. This lag is due to the inherent complexity of plants as multicellular organisms with longer life cycles, tissue-specific gene expression, limitations on reproducible transgene introduction (Liu et al., Reference Liu, Panda, Edwards, Swanson, Yi, Pandesha, Hung, Klaas, Ye, Collins, Renken, Gilbertson, Veena, Hancock and Slotkin2024) and susceptibility to gene silencing (Stam, Reference Stam1997), all of which complicate predictive modelling. Furthermore, plants are exposed to diverse external signals, and spatial-temporal variations in gene expression add further challenges.
Artificial intelligence (AI) approaches could offer new solutions to accurately predict circuit performance in plants and improve the slow DBTL cycle in plant circuit development, particularly when coupled to large-scale genomic, transcriptomic and environmental data. These models can handle non-linear interactions within complex regulatory networks and predict behaviour across tissues and developmental stages (Rai et al., Reference Rai, Wang, O’Connell, Patel and Bashor2024). By automating design processes and leveraging high-throughput testing, AI could significantly accelerate the development of plant circuits, overcoming the unique challenges of plant synthetic biology.
3.4. Implementing circuits in crops
To date, synthetic gene circuits have been successfully implemented in model plants such as Arabidopsis and N. benthamiana. It will be crucial to test and implement these circuits in agriculturally important crop species to address challenges related to biotic and abiotic stresses, growth and yield.
The implementation of synthetic gene circuits in crops will require the identification and testing of species-specific circuit components, for example, for the design of the sensors and integrators. To date, the availability of such parts is limited in both model and crop plants. The increasing application of single cell transcriptome and chromatin accessibility profiling of crop species will greatly advance the identification of cell-type-specific promoters and regulatory regions (He et al., Reference He, Luo, Zhou, Zhu, Lan and Chen2024; Marand et al., Reference Marand, Chen, Gallavotti and Schmitz2021). This will underpin development of a wide range of different input components for implementing synthetic gene circuits in vivo.
Several additional challenges may hinder the implementation of synthetic circuits in crops. These include the difficulty and time required to generate stable transgenic lines, testing the functionality of circuits over a few generations and ensuring the stability of constructs while avoiding gene silencing. The generation of more accurate models of circuit performance, for example through computational models trained on high-throughput cell-type-specific genomics and circuit testing datasets, could reduce the burden of these inherently challenging crop engineering steps by more accurate prediction of circuit designs likely to function as desired in vivo. Addressing these key challenges will significantly advance the field of plant synthetic biology and enable the development of crop plants with novel traits to address specific agricultural challenges.
4. Concluding remarks
Recent years have seen the successful development and implementation of a range of synthetic gene circuits in plants that advance our capabilities to achieve programmable control of gene expression. These circuits are capable of retaining a memory of the past stimuli and can dynamically switch between ‘ON’ and ‘OFF’ states, depending on the technology used for their construction. Though these circuits function in a predictable manner in model plants such as Arabidopsis and N. benthamiana, multiple major challenges remain in their further advancement, optimisation, and implementation, which will require focused experimental and computational developments. These include the need for high-throughput circuit synthesis and testing, coupled with data-rich multi-component quantitative assessment of circuit activity, to generate the data that will underpin accurate circuit modelling, which is crucial for further and faster advancements.
Synthetic gene circuits will provide completely new capabilities for engineering crops to address agriculture challenges and introduce new agronomic traits. However, the current circuit designs optimised for model plants may not function in a predictable manner in different crop species. Therefore, it will be crucial to identify effective biological parts for each species, and implement high-throughput circuit synthesis and quantitative assessment frameworks to accelerate successful circuit function and adoption in diverse crops. These advancements will help us to introduce innovative agronomic traits in crops, thus enabling sustainable food production to meet the growing global demand.
Acknowledgements
We thank Quantitative Plant Biology for inviting us to submit this review article. Figures were generated using BioRender.com.
Competing interest
The authors declare no competing interests.
Data availability statement
Data and code availability is not applicable as no new data were generated or created.
Author contributions
A.K. and R.L. wrote the manuscript.
Funding statement
This work was supported by the following grants to R.L.: Australian Research Council (ARC) Center of Excellence in Plants for Space (CE230100015), ARC Discovery Project DP240103385, and NHMRC Investigator Grant GNT1178460.
Open peer review
To view the open peer review materials for this article, please visit http://doi.org/10.1017/qpb.2025.3.







Comments
No accompanying comment.