Introduction
The field of drug discovery and design is one of the most vibrant areas of research, with many groups from academic and industrial settings contributing to its advancement. The identification of small molecules binding to protein or RNA targets, and the related structural and functional information is of fundamental importance to modern drug discovery. One way to identify small molecule binders is high throughput screening (HTS), where millions of compounds are tested for the desired biological activity (Martis et al., Reference Martis, Radhakrishnan and Badve2011). However, HTS is time-consuming and expensive (DiMasi et al., Reference Dimasi, Hansen and Grabowski2003; Workman, Reference Workman2003; Hopkins, Reference Hopkins2009). An alternative to HTS is computer-aided drug design (CADD) or virtual HTS (Leelananda and Lindert, Reference Leelananda and Lindert2016). CADD provides the capability of screening millions of compounds virtually. This significantly reduces the number of molecules that need to be tested biochemically. Therefore, this approach can cut cost and accelerate the preliminary stage of drug development. CADD has been successfully applied to numerous disease-related target systems advancing research in treatments for congestive heart failure (Lindert et al., Reference Lindert, Li, Sykes and Mccammon2015a; Cai et al., Reference Cai, Li, Pineda-Sanabria, Gelozia, Lindert, West, Sykes and Hwang2016; Aprahamian et al., Reference Aprahamian, Tikunova, Price, Cuesta, Davis and Lindert2017; Coldren et al., Reference Coldren, Tikunova, Davis and Lindert2020), several types of cancer (Lee et al., Reference Lee, Jeong, Lee, Song, Kim, Lee and Kim2010; Ravindranathan et al., Reference Ravindranathan, Mandiyan, Ekkati, Bae, Schlessinger and Jorgensen2010; Chuang et al., Reference Chuang, Cheng, Leu, Chuang, Tzou and Chen2015; Lee et al., Reference Lee, Lin, Wen, Wang and Lin2019; Fratev et al., Reference Fratev, Gutierrez, Aguilera, Tyagi, Damodaran and Sirimulla2021; Hantz and Lindert, Reference Hantz and Lindert2022), and various infectious diseases (Durrant et al., Reference Durrant, Cao, Gorfe, Zhu, Li, Sankovsky, Oldfield and Mccammon2011; Zhu et al., Reference Zhu, Zhang, Sinko, Hensler, Olson, Molohon, Lindert, Cao, Li, Wang, Wang, Liu, Sankovsky, De Oliveira, Mitchell, Nizet, Mccammon and Oldfield2013; Sinko et al., Reference Sinko, Wang, Zhu, Zhang, Feixas, Cox, Mitchell, Oldfield and Mccammon2014; Lindert et al., Reference Lindert, Tallorin, Nguyen, Burkart and Mccammon2015b; de Sousa et al., Reference De Sousa, Combrinck, Maepa and Egan2020).
In practice, commonly utilised CADD approaches may vary depending on the availability and type of experimental information. The two main methods being employed are structure-based drug design (SBDD) and ligand-based drug design (LBDD). In the structure-based approach, the 3D structures of targets are known. They are generally elucidated using techniques such as X-Ray crystallography, nuclear magnetic resonance spectroscopy and cryogenic electron microscopy and can be further supplemented with molecular dynamic simulations (Salsbury, Reference Salsbury2010), computational structure prediction (Dorn et al., Reference Dorn, Silva, Buriol and Lamb2014; Seffernick and Lindert, Reference Seffernick and Lindert2020) and design (Huang et al., Reference Huang, Boyken and Baker2016) methods. These structures are then used for generating or screening potential small molecule ligands by predicting interactions with the target. On the other hand, ligand-based approaches are utilised when the 3D structure of the target is unknown. However, LBDD relies on the existence of a large set of ligands whose potency against a biological target of interest is known. Using such information, a correlation between the structures and properties of known ligands and their experimentally determined biological activities can be derived. This structure–activity relationship may in turn be used to design new drugs. Over the last decade, there has been a rapid shift in the field of drug development aimed at improving CADD approaches using a variety of machine learning (ML) techniques (Cao et al., Reference Cao, Liu, Tan, Song, Shu, Li, Zhou, Bo and Xie2018). Recently, others have reviewed the general role of artificial intelligence throughout the drug discovery pipeline (Hessler and Baringhaus, Reference Hessler and Baringhaus2018; Patel et al., Reference Patel, Shukla, Huang, Ussery and Wang2020; Carracedo-Reboredo et al., Reference Carracedo-Reboredo, Liñares-Blanco, Rodríguez-Fernández, Cedrón, Novoa, Carballal, Maojo, Pazos and Fernandez-Lozano2021; Gallego et al., Reference Gallego, Naveiro, Roca, Ríos Insua and Campillo2021; Paul et al., Reference Paul, Sanap, Shenoy, Kalyane, Kalia and Tekade2021; Dara et al., Reference Dara, Dhamercherla, Jadav, Babu and Ahsan2022). Notably, in this work, we provide an overview of recent applications of deep learning (DL) in CADD methods, with a specific focus on SBDD.
Structure-based drug design
SBDD is the most commonly utilised approach when the three-dimensional structure of the drug target is known. In SBDD, one first identifies the structural data of the target either experimentally or through computational modelling. Next, a docking algorithm is used to position compounds from a database into the selected region of the target. These compounds are then scored and ranked using a score function and top hits are tested experimentally. Over the past few years, SBDD methodologies have gone through significant improvements in speed and accuracy. The introduction of ML techniques to SBDD approaches has also enhanced performance (Cao et al., Reference Cao, Liu, Tan, Song, Shu, Li, Zhou, Bo and Xie2018). DL techniques, a subdivision of ML with multi-layered neural network architectures, are increasingly utilised in drug discovery to capture complex data patterns and predict outcomes (Schmidhuber, Reference Schmidhuber2015). Reinforcement learning (RL) is yet another subdivision of ML, where a computer model is trained by rewarding desired outcomes and penalised for undesired ones (Botvinick et al., Reference Botvinick, Ritter, Wang, Kurth-Nelson, Blundell and Hassabis2019). The combination of DL and RL techniques (DRL) in the field of drug design has also been revolutionary and has led to several breakthroughs in recent years (Olivecrona et al., Reference Olivecrona, Blaschke, Engkvist and Chen2017; Neil et al., Reference Neil, Segler, Guasch, Ahmed, Plumbley, Sellwood and Brown2018; Popova et al., Reference Popova, Isayev and Tropsha2018; Ståhl et al., Reference Ståhl, Falkman, Karlsson, Mathiason and Boström2019; Zhou et al., Reference Zhou, Kearnes, Li, Zare and Riley2019). Applications of these techniques in SBDD can be categorised into three main areas of focus: de novo drug design, binding site prediction, and binding affinity prediction.
De novo drug design
De novo drug design refers to the exploration of a broad chemical space and creation of new chemical compounds without the need for a starting template and was first introduced by Danziger and Dean (Reference Danziger and Dean1989). The chemical search space for identifying novel compounds is virtually infinite and sufficiently sampling this space is a primary challenge in de novo drug design. In order to resolve this issue, a set of restraints is typically incorporated. For instance, the physical and chemical properties of a target’s active site are key constraints for ligand design. Structural and chemical information plays an immense role in reducing the search space to prevent the sampling of generations of compounds that might be synthetically unobtainable. Two sampling methods, atom-based and fragment-based, are primarily used for compound generation in de novo drug design (Gillet et al., Reference Gillet, Johnson, Mata, Sike and Williams1993; Wong et al., Reference Wong, Luo and Chan2011; Schneider and Fechner, Reference Schneider and Fechner2022). In atom-based sampling (Fig. 1a), an initial atom is used as a seed inside the active site of the target. From this seed, a diverse set of compounds are grown by varying the number of atoms and hybridization states of each atom (Nishibata and Itai, Reference Patel, Shukla, Huang, Ussery and Wang1991). Compounds generated from atom-based sampling have high structural diversity. However, the computational cost in narrowing down the lead compound with the atom-based method can increase exponentially with the size of the compound being designed (Hartenfeller and Schneider, Reference Hartenfeller and Schneider2011). Alternatively, fragment-based sampling (Fig. 1b) circumvents this issue by exploring a database of fragments. These fragments are then used as a seed in the active site to build the rest of the compound. This approach significantly narrows the chemical search space while maintaining good structural diversity (Böhm, Reference Böhm1992; Gillet et al., Reference Gillet, Johnson, Mata, Sike and Williams1993; Pearlman and Murcko, Reference Pearlman and Murcko1996; Clark et al., Reference Clark, Frenkel, Levy, Li, Murray, Robson, Waszkowycz and Westhead2022).

Fig. 1. Illustration of computational de novo drug design. (a) In atom-based drug design, the small molecule is built atom by atom by sampling additions of many different types of atoms. (b) In fragment-based drug design, the small molecule is built by sampling additions of a library of fragments.
DL and deep reinforcement learning techniques have been used in numerous cases to improve the performance of de novo drug design. Common DL architectures utilised in de novo drug design include recurrent neural network (RNN), graph neural network (GNN), and graph convolutional neural network (GCNN). RNNs make use of sequential data or time series data and are known for their internal memory which takes information from prior inputs to influence the current input and output. In contrast, traditional deep neural networks assume that inputs and outputs are independent of one another (Dupond, Reference Dupond2019). RNNs have been recently expanded with long short-term memory (LSTM) networks. Introduced by Hochreiter and Schmidhuber (Reference Hochreiter and Schmidhuber1997), LSTM are used as the building blocks for the layers of a RNN and enable RNNs to remember inputs over a long period of time. This memory is commonly referred to as a gated cell, meaning the cell decides whether to store or delete information based on the importance or weight it assigns to the information. GNNs are a class of artificial neural networks used for processing data that can be depicted as graph structures. GNNs operate on an information diffusion mechanism, meaning a graph is processed by a set of units which are linked according to the graph connectivity. The units exchange information in a pairwise fashion until a stable equilibrium is reached (Scarselli et al., Reference Scarselli, Gori, Tsoi, Hagenbuchner and Monfardini2009). This behaviour is similar to an RNN as weights are shared in each recurrent step. In contrast to RNNs and GNNs, GCNNs do not share weights between hidden layers. GCNNs generalise classical convolutional neural networks (CNNs) to graph-structured data. In this representation, structural information can be incorporated to model connections among entities and create further insights in the data. GCNNs can utilise the graph structure and aggregate node information from the neighbourhoods in a convolutional fashion (Zhang et al., Reference Zhang, Tong, Xu and Maciejewski2019). Deep reinforcement learning (DRL) combines artificial neural networks with a framework of reinforcement learning (RL) that helps agents (i.e. the algorithm making decisions/actions) learn how to reach their goals. RL considers the problem of an agent learning to make decision based on trial-and-error from rewards or punishments. DRL incorporates DL by allowing the agent to make decisions from unstructured data or domain heuristics. We will review several notable recent examples of these architectures below.
Jeon and Kim (Reference Jeon and Kim2020) created an atom-based de novo method to design novel compounds named Molecule Optimization by Reinforcement Learning and Docking (MORLD). MORLD is a deep generative model that uses binding affinities, obtained from docking simulations, as rewards in the reinforcement learning. In MORLD, a compound is optimised by Molecule Deep Q-Networks (MolDQN) (Zhou et al., Reference Zhou, Kearnes, Li, Zare and Riley2019), based on reinforcement learning and chemistry domain knowledge. Compounds are optimised in single action steps, meaning either an addition or removal of an atom or bond in a chemically valid manner. This is controlled by the user as atom types for modification need to be specified. The modified molecule is then evaluated with different score functions such as the synthetic accessibility (SA) (Ertl and Schuffenhauer, Reference Ertl and Schuffenhauer2009) and quantitative estimate of drug-likeness (QED) (Bickerton et al., Reference Bickerton, Paolini, Besnard, Muresan and Hopkins2012). The SA score is a metric that is used to estimate the ease of synthesis of drug-like molecules. SA score ranges from 1 (easy to synthesise) to 10 (difficult to synthesise). QED provides a quantitative metric for assessing druglikeness of target (and/or predicted) compounds. This value ranges between 0 and 1. A compound QED score of 0 indicates that all properties of that compound are unfavourable and vice-versa. These scores are weighted and used to guide compound modification with MolDQN in the next state. This process is repeated until the compound reaches a terminal state. In the terminal state, the modified compound is docked to the target with QuickVina2 (Alhossary et al., Reference Alhossary, Handoko, Mu and Kwoh2015). The resulting docking score is given to MolDQN as a reward and a compound is generated. This is repeated until a specified number of training episodes are reached. MORLD was shown to significantly decrease the time needed to design a novel inhibitor for the target discoidin domain receptor 1 kinase (DDR1) compared to the deep generative model GENTRL (Zhavoronkov et al., Reference Zhavoronkov, Ivanenkov, Aliper, Veselov, Aladinskiy, Aladinskaya, Terentiev, Polykovskiy, Kuznetsov, Asadulaev, Volkov, Zholus, Shayakhmetov, Zhebrak, Minaeva, Zagribelnyy, Lee, Soll, Madge, Xing, Guo and Aspuru-Guzik2019). GENTRL took approximately 46 days (with training data) to successfully design a novel inhibitor against DDR1, whereas MORLD achieved the same result in less than 2 days without any training data. MORLD was also successfully able to generate agonists for D4 dopamine receptor (D4DR) without any initial lead compound information. MORLD can be used in situations where the lead compound is available, or when the lead compound must be identified from virtual screening or from scratch. MORLD has a free web server and source code open to the public. While MORLD is a successful tool for drug design, it has a few limitations: (a) the MORLD algorithm is trained on Q-value. Q-value is the expected future rewards, that is a p-value that has been adjusted for the false discovery rate. For example, a q-value of 2% means that 2% of significant results will result in false positives. This limits the diversification of optimised compounds, (b) as an atom-based model it is unable to explore a large chemical space, and (c) the SA and QED scores are not perfect. Therefore, generated compounds are sometimes chemically inaccurate. Additionally, MORLD is also not a suitable method for drug design when the target protein is disordered or does not have a druggable binding pocket. While a relatively new program, MORLD has been cited in many studies to acknowledge the usefulness of DRL in the field of drug discovery. To date, this algorithm has been applied to only the DDR1 system as mentioned previously.
Ma et al. (Reference Ma, Terayama, Matsumoto, Isaka, Sasakura, Iwata, Araki and Okuno2021) created the Structure-Based de Novo Molecular Generator (SBMolGen), another DL method for de novo drug design based on Monte Carlo tree search (MCTS) (Browne et al., Reference Browne, Powley, Whitehouse, Lucas, Cowling, Rohlfshagen, Tavener, Perez, Samothrakis and Colton2012a, Reference Browne, Powley, Whitehouse, Lucas, Cowling, Rohlfshagen, Tavener, Perez, Samothrakis and Colton2012b) and docking simulations. Molecular generation in SBMolGen is done by ChemTS (Yang et al., Reference Yang, Zhang, Yoshizoe, Terayama and Tsuda2017), and rDock (Ruiz-Carmona et al., Reference Ruiz-Carmona, Alvarez-Garcia, Foloppe, Garmendia-Doval, Juhos, Schmidtke, Barril, Hubbard and Morley2014) is employed for docking generated compounds to the target. In the SBMolGen methodology, ChemTS first generates compounds with MCTS and a recurrent neural network (RNN) (Kaur and Mohta, Reference Kaur and Mohta2019). These generated models are then docked using rDock and evaluated. Based on the top scoring model from the docking simulation, SBMolGen reweights the search tree in ChemTS. Additional molecules are then generated by ChemTS with the new weights. By repeating this process with a RNN, SBMolGen is able to generate compounds that are aware of the target it is binding to. The RNN model in SBMolGen was trained on 250,000 molecules from the ZINC database (Irwin et al., Reference Irwin, Sterling, Mysinger, Bolstad and Coleman2012). The results of SBMolGen were benchmarked with four target proteins: cyclin-dependent kinase 2 (CDK2), epidermal growth factor receptor erbB1 (EGFR), adenosine A2A receptor (AA2AR), and beta-2 adrenergic receptor (ADRB2). For each target, the generated molecules were evaluated against known actives from the Database of Useful Decoys: Enhanced (DUD-E) (Mysinger et al., Reference Mysinger, Carchia, Irwin and Shoichet2012). Molecules generated with SBMolGen covered a larger chemical space than the ZINC data set, had an SA score of less than 3.5 and generally many of these compounds had QED scores greater than 0.8. Furthermore, SBMolGen was also able to find compounds that had a better docking score (as compared to known compounds) for all four target proteins. Fragment molecular orbital (FMO) (Kitaura et al., Reference Kitaura, Ikeo, Asada, Nakano and Uebayasi1999) analysis of the target protein and generated molecules suggested that SBMolGen designed molecules with high binding affinity. Benchmark results of SBMolGen were compared against GENTRL and showed that SBMolGen was able to produce molecules that had better docking scores. While SBMolGen is faster than GENTRL, it is still limited by computation time. In addition, docking scores of SBMolGen were based on SA score, Lipinski’s rule of five (Lipinski et al., Reference Lipinski, Lombardo, Dominy and Feeney2001) and PubChem (Kim et al., Reference Kim, Chen, Cheng, Gindulyte, He, He, Li, Shoemaker, Thiessen, Yu, Zaslavsky, Zhang and Bolton2021) filters. While these are useful filters, comprehensive drug discovery still requires the optimization of a large number of molecular properties (Winter et al., Reference Winter, Montanari, Steffen, Briem, Noé and Clevert2019). SBMolGen has been employed as benchmark in several other method development studies.
Li et al. (Reference Li, Pei and Lai2021b) created DeepLigBuilder, another atom-based DL method for de novo drug design. The deep generative model in DeepLigBuilder is able to design and optimise the 3D structures of ligands directly inside the binding pocket of the target. This method combines a graph convolutional neural network (L-Net) and MCTS, a common technique in reinforcement learning. L-Net iteratively generates and refines molecules by using a state encoder and policy network. During this stage, the state encoder analyses the state of the molecules and passes it to the policy network. The policy network then decides the type, bond order, coordinates, and the number of new atoms to be added. This combination allows the DeepLigBuilder to generate molecules with high binding affinity within the binding pocket. Benchmarking results indicated that L-Net had an overall validity of 96% compared to G-SchNet (Gebauer et al., Reference Gebauer, Gastegger, Hessmann, Müller and Schütt2022) which had an output validity of only 77%. The average RMSD of the generated compounds to optimised structures of the generated models with MMFF94 (Halgren, Reference Halgren1996) was only about 0.61 Å. The QED score was above 0.5 for 83.9% of the generated compounds and it was possible to correctly predict the overall distribution of molecular properties. L-Net was also able to re-create the chemical space of the training set almost identically. While L-Net’s performance was highly remarkable, it struggled to generate ring structures as observed by the high standard deviations for bond lengths and torsion angles for aromatic rings. DeepLigBuilder was also tested to generate 3D structures of compounds (as well as lead optimization of known inhibitors) inside the binding pocket of SARS-CoV-2 main protease. The MCTS (as opposed to random search) in DeepLigBuilder successfully lowered the smina scores (Koes et al., Reference Koes, Baumgartner and Camacho2013) during lead optimization. Furthermore, DeepLigBuilder was able to generate compounds for SARS-CoV-2 main protease with novel chemical structures and high predicted binding affinity and had a success rate of 78.1%. DeepLigBuilder has been cited in several other studies for its use in generating lead compounds for SARS-CoV-2 as discussed above. However, to date, this algorithm has not been applied to other systems.
DeepScaffold is another DL tool for scaffold-based (can also be thought of as fragment-based) drug discovery created by Li et al. (Reference Li, Hu, Wang, Zhou, Zhang and Liu2020). DeepScaffold is able to design compounds from cyclic skeletons, classical molecular scaffolds, and pharmacophore queries. DeepScaffold uses three different methods to enable these different inputs. When the input is a cyclic skeleton, DeepScaffold uses a graph convolutional neural network and variational autoencoder (VAE) (Kingma and Welling, Reference Kingma and Welling2014) to complete atom and bond types and generate a scaffold. However, if the input is a molecular scaffold, then the program uses a generative model (Li et al., Reference Li, Zhang and Liu2018), from the authors’ previous study, to build the side chains. DeepScaffold can also filter out generated molecules that do not match the input pharmacophore query. DeepScaffold was trained on a dataset that contained 914,464 molecules from the ChEMBL databases with QED larger than 0.5. DeepScaffold was then evaluated for chemical validity and diversity, distribution of molecular properties and side chains. The performance of DeepScaffold to produce chemically valid and unique scaffolds was remarkable with an average of 98.9 and 69.1%, respectively. Molecular properties (molecular weight, solubility, and QED) of the test set molecules against the generated molecules indicate good correlation for scaffolds with more atoms. The model also demonstrated a diversified substitution pattern of side chains and was biased towards adding more atoms to nitrogen atoms. Finally, DeepScaffold model was tested on GPCRs. The model was able to produce chemically valid molecules, however, performed rather poorly to produce unique molecules with increasing structural complexity of scaffolds. The model was also able to reproduce known actives for the GPCRs reasonably well. DeepScaffold has been mentioned in several studies. This algorithm has not yet been applied to other target systems. However, it was referenced for successful scaffold hopping during compound design (Zhang et al., Reference Zhang, Liu, Fan, Leung, Yao and Liu2022). Additionally, DeepScaffold has one of the highest percentages for validity of generated compounds as compared to other scaffold-based de novo methods (Joshi et al., Reference Joshi, Gebauer, Bontha, Khazaieli, James, Brown and Kumar2021).
MolAICal, created by Bai et al. (Reference Bai, Tan, Xu, Liu, Huang and Yao2021), is another fragment-based drug design algorithm. The two modules in MolAICal are trained with a DL model on fragments from the food and drug administrations (FDA) approved drugs and drug-like ligands from the ZINC database. MolAICal can directly generate 3D structures of drug-like compounds in the protein pockets. It employs a sequence-based generative model and a GNN for fragment generation and the fragment is then grown stochastically. A total of 21,064 fragments of FDA-approved drugs and 1,060,000 drug-like ligands from the ZINC database were used to train the generative model. When the grown ligand reaches a certain length, a genetic algorithm is employed to optimise the ligand into the protein pocket based on the ligand-receptor binding score. The docking score used by MolAICal is the Vinardo score (Quiroga and Villarreal, Reference Quiroga and Villarreal2016) from AutoDock Vina (Trott and Olson, Reference Trott and Olson2010). The Vinardo score is a summation of several energetic terms. These are Gaussian steric attractions, quadratic steric repulsions, Lennard-Jones potentials, electrostatic interactions, hydrophobic interactions, non-hydrophobic interactions, and non-directional hydrogen bonds. Vinardo scores are reported as the Gibbs free energy (ΔG) of binding in the unit of kcal mol−1. Therefore lower Vinardo scores are more favourable. However, in MolAICal, the coefficients of the Vinardo score were optimised based on a set of 2,903 protein-ligand complexes with experimental data from the PDBbind database (Wang et al., Reference Wang, Fang, Lu, Yang and Wang2005). MolAICal was then tested to design drugs for glucagon receptor (GCGR) and SARS-CoV-2 main protease. For both the GCGR and the SARS-CoV-2 main protease, MolAICal was able to generate compounds that were diversified and had good theoretical binding affinity. In a recent study, MolAICal was employed to calculate the molecular synthetic accessibility score of traditional Chinese medicines in attempts to inhibit the SARS-CoV-2 nucleocapsid protein (Chen et al., Reference Chen, Wei, Qin, Hou, Zang, Zhang and Chen2022). In another application, MolAICal was utilised to calculate the relative binding energy of compounds from the ZINC database to inhibit carbonic anhydrase as a treatment for altitude sickness (Ali et al., Reference Arús-Pous, Patronov, Bjerrum, Tyrchan, Reymond, Chen and Engkvist2022).
Finally, Arús-Pous and co-authors describe a fragment-based method that employs DL architecture to generate scaffolds based on SMILES string (Arús-Pous et al., Reference Arús-Pous, Patronov, Bjerrum, Tyrchan, Reymond, Chen and Engkvist2020). In this drug-design architecture, a scaffold is generated through an RNN, that is composed of three LSTM cells (Hochreiter and Schmidhuber, Reference Hochreiter and Schmidhuber1997), from a SMILES string. Next, the scaffold is ‘decorated’ with side-chains at each attachment point in the scaffold exhaustively. The generated molecules are then filtered based on drug-like properties such as molecular weight, QED, SA, and/or user-specified properties. The decorator model was trained on a set of 4,211 Dopamine Receptor D2 active modulators. Following the training phase, the authors evaluated their generative model and the model was able to produce a diverse set of scaffolds and obtain a large amount of actives. The architecture implemented by Arús-Pous and co-authors is extremely versatile and can be combined with molecular generative architectures. This work by Arús-Pous and co-authors has been highlighted primarily in reviews of artificial intelligence in drug design.
A brief summary of all the previously mentioned methods, along with the respective links to the freely accessible code or webserver, can be found in Table 1. The methods mentioned here capture many of the key areas of how DL has influenced de novo drug design. Nonetheless, this is still an active area of research with novel applications of DL algorithms continuously being developed.
Table 1. Summary of de novo drug design methods

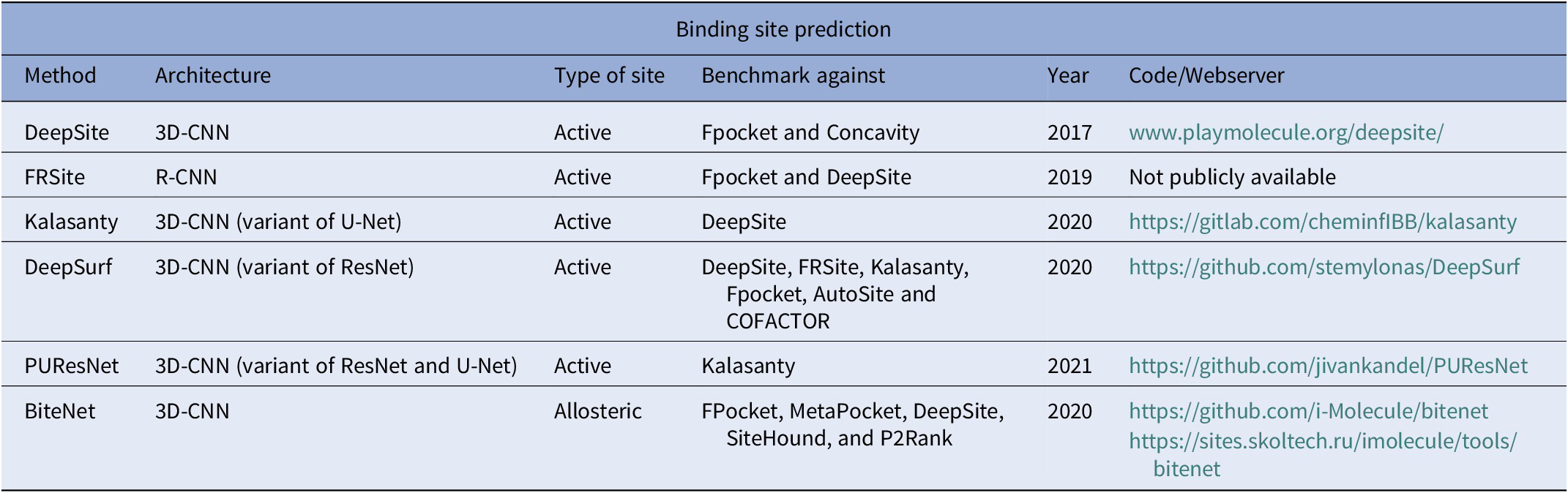
Binding site prediction
Ligand binding sites in proteins (Fig. 2) encode necessary chemical and structural information for the successful execution of SBDD. Correct identification of these binding sites may help in the understanding of protein function and aid in the design of better drug-like small molecules. While these functional sites can be experimentally determined, the time and cost required to perform such experiments are quite significant. In contrast, computational prediction of protein-ligand binding sites is relatively inexpensive and significantly faster, thus expediting the drug development process. Several computational methods have been proposed over the years for structure-based binding site prediction (Huang, Reference Huang2009; Le Guilloux et al., Reference Le Guilloux, Schmidtke and Tuffery2009; Yu et al., Reference Yu, Zhou, Tanaka and Yao2010; Tsujikawa et al., Reference Tsujikawa, Sato, Wei, Saad, Sumikoshi, Nakamura, Terada and Shimizu2016; Dias et al., Reference Dias, Nguyen, Jorge and Gomes2017; Wu et al., Reference Wu, Peng, Zhang and Yang2018). These methods can be roughly categorised as geometry-based, energy-based, and templated-based (Mylonas et al., Reference Mylonas, Axenopoulos and Daras2021). Geometry-based methods analyse the geometry of the molecular surface of a target protein in order to locate surface cavities. Common approaches to find surface cavities within geometry-based methods include grid system scanning (Weisel et al., Reference Weisel, Proschak and Schneider2007), probe sphere filling (Yu et al., Reference Yu, Zhou, Tanaka and Yao2010) and alpha spheres (Le Guilloux et al., Reference Le Guilloux, Schmidtke and Tuffery2009). Energy-based binding site prediction methods involve searching for energetically favourable interactions between protein atoms and a chemical probe with the aid of a force field (Kozakov et al., Reference Kozakov, Grove, Hall, Bohnuud, Mottarella, Luo, Xia, Beglov and Vajda2015; Tsujikawa et al., Reference Tsujikawa, Sato, Wei, Saad, Sumikoshi, Nakamura, Terada and Shimizu2016). Schrodinger’s SiteMap (Halgren, Reference Halgren2007, Reference Halgren2009) program combines geometry and energy-based properties to identify potential protein–ligand and protein–protein binding sites. Finally, template-based binding site prediction relies on the assumption that proteins that share structural similarity to the target protein also share functional similarity (Dey et al., Reference Dey, Cliff Zhang, Petrey and Honig2013). One of the first ML-based binding site prediction algorithms was CryptoSite (Cimermancic et al., Reference Cimermancic, Weinkam, Rettenmaier, Bichmann, Keedy, Woldeyes, Schneidman-Duhovny, Demerdash, Mitchell, Wells, Fraser and Sali2016). CryptoSite uses as support vector machine (SVM) to classify residues belonging to a binding site on a score range of 0 (not likely)–1 (most likely). Efforts in DL-based binding site prediction methods are fairly new and date back to as early as 2017.

Fig. 2. Illustration of computational binding site prediction. In this methodology, 3D voxels are used to identify regions of the protein as potential binding sites (shown as yellow rectangles in the figure). Next, these sites are ranked from most to least probable for a ligand to bind.
Convolutional neural networks (CNNs) are a popular DL architecture for identifying potential binding sites. CNNs are regularised versions of fully connected networks, meaning each neuron in one layer is connected to all neurons in the next layer (Albawi et al., Reference Albawi, Mohammed and Al-Zawi2017). In a 2D-CNN, the kernel or filter used to extract features from images moves in two directions (x and y) and the input/output data is three-dimensional. 2D-CNNs are typically used on image data. A region-based CNN (R-CNN), is a type of 2D-CNN that implements a selective search algorithm that generates region proposals that are then provided to the CNN architecture to compute the features (Girshick et al., Reference Girshick, Donahue, Darrell and Malik2014). Region proposals are small regions of the image that potentially contain objects of interest in the input image. The benefit of this method is that the algorithm generates approximately 2,000 region proposals which greatly reduces the computational cost compared to computing CNN features for the entire input image. In a 3D-CNN, the kernel moves in three directions (x, y, and z) and the input/output of the model are four-dimensional. These algorithms are typically used on three-dimensional image data (Hedegaard et al., Reference Hedegaard, Heidari, Iosifidis, Iosifidis and Tefas2022). A popular 3D-CNN architecture is U-Net (Ronneberger et al., Reference Ronneberger, Fischer and Brox2015) which has been shown to outperform traditional sliding window convolutional networks. The model provides many advantages over patch-based segmentation approaches as it preserves the full context of the input image through an end-to-end pipeline process for the entire image and requires few training samples. One increasingly popular CNN architecture is ResNet (He et al., Reference He, Zhang, Ren and Sun2016). ResNet overcomes the vanishing gradient problem, which occurs when the neural network training algorithm attempts to find weights that bring the loss function to a minimal value. If the network has too many layers, the gradient becomes increasingly small until it disappears and optimization cannot continue. ResNet overcomes this problem by creating a deep residual learning framework. This framework consists of shortcut connections that add intermediate inputs to the output of a group of convolution blocks. The shortcut connections perform identity mapping, allowing for smoother gradient flow and ensures important features are carried until the final layers. We review several notable recent examples of these architectures below.
Jiménez et al. (Reference Jiménez, Doerr, Martínez-Rosell, Rose and De Fabritiis2017) developed the first DL-based protein-ligand binding site prediction algorithm, DeepSite. Based on a 4-layer convolutional neural network (DCNN) architecture, DeepSite treats the target protein as an image on a 3D grid. Each atom within the voxels of the grid is defined by atom-based physicochemical properties obtained from AutoDock 4 (Morris et al., Reference Morris, Huey, Lindstrom, Sanner, Belew, Goodsell and Olson2009). Next, the entire grid is divided into subgrids in order to defined the local properties of smaller protein areas. These subgrids are then scored by the DCNN as potential sites for ligand binding. The DCNN architecture in DeepSite was trained on 7,622 proteins from the sc-PDB (Desaphy et al., Reference Desaphy, Bret, Rognan and Kellenberger2015) database. Pocket prediction performance was evaluated by distance and shape of the predicted binding site to that of the real binding site. To accomplish this, the authors make use of two metrics, distance to centre of binding site (DCC) and discretized volumetric overlap (DVO). These metrics were also used to benchmark DeepSite against Fpocket (Le Guilloux et al., Reference Le Guilloux, Schmidtke and Tuffery2009) and Concavity (Capra et al., Reference Capra, Laskowski, Thornton, Singh and Funkhouser2009) (binding site prediction algorithms). Based on the binding site analysis metrics, DeepSite was able to provide a more accurate binding site prediction compared to Fpocket and Concavity. Further analysis of DeepSite on proteins with diverse folds in the SCOPe database showed no bias towards any specific protein fold. However, there is still room for improvement as DeepSite does not have descriptors for atom-based polarizability effects or descriptors for water molecules. Since its release, DeepSite has been a highly successful tool and has been referenced in several studies. A few notable applications are its use to study the binding process and interaction of antibiotic drugs and lysozymes (Rial et al., Reference Rial, González-Durruthy, Liu and Ruso2022), analysis of the potential interaction mechanism of sweeteners with aroma compounds in passion fruit juice (Xiao et al., Reference Xiao, Jiang and Niu2022), and identifying binding pockets for drug candidates for Hepatocellular carcinoma (predominant subtype of liver cancer) (Tang et al., Reference Tang, Guo, Yang, Wang, Zhang and Wang2022).
FRSite, introduced by Jiang et al. (Reference Jiang, Wei, Zhang, Wang, Wang and Li2019), is another binding site prediction method based on a region-based convolutional neural network (R-CNN), an object-detection DL architecture (Ren et al., Reference Ren, He, Girshick, Sun, Cortes, Lawrence, Lee, Sugiyama and Garnett2015). In this methodology, the target protein is treated as a 3D image and the binding site is treated as the object within the image to be detected. Similar to DeepSite, FRSite also treats each atom in the target protein on a grid with eight descriptors from high-throughput molecular dynamics (HTMD) (Doerr et al., Reference Doerr, Harvey, Noé and De Fabritiis2016). Next, the entire grid is fed into a region proposal network-3D (RPN-3D) module in FRSite to detect potential binding sites. Once these potential binding sites are found by RPN-3D, FRSite uses its classifier to score the binding site. FRSite was trained on the same training dataset as DeepSite. Following the training, FRSite was evaluated for DCC and DVO as well as compared to those of other prediction methods. Compared to Fpocket and DeepSite, FRSite outperformed the other algorithms in predicting more accurate binding sites of proteins. Additionally, FRSite is also able to predict the receptor grid of the binding site (which can be used to estimate the size of the pocket). The authors further showed that the DVO estimated by FRSite was more accurate than that of Fpocket. FRSite has been referenced in several review articles and as benchmark in other method development studies.
Stepniewska-Dziubinska et al. (Reference Stepniewska-Dziubinska, Zielenkiewicz and Siedlecki2020) introduced another DL-based binding site prediction method called Kalasanty. Similar to DeepSite, Kalasanty also treats the target protein as a 3D image. However, Kalasanty uses a DL model that is more similar to U-Net (Ronneberger et al., Reference Ronneberger, Fischer and Brox2015) to treat the problem as a 3D image segmentation problem. In this approach, each featurized segment (by atomic properties) of the protein is assigned a certain probability of being a part of a pocket where a small molecule can dock. The DL model in Kalasanty was trained on 15,860 structures with binding site data from sc-PDB dataset. Prediction results from Kalasanty were then evaluated for correct DCC and DVO as well compared to those from DeepSite. Kalasanty correctly predicted binding sites two times better than DeepSite. Furthermore, Kalasanty had DCC values lower (i.e. closer to centre of the actual binding site) than DeepSite. Additionally, the binding sites predicted by DeepSite were bigger (lower DVO) and not accurately modelled when compared to that of Kalasanty (higher DVO). While Kalasanty is a successful binding site prediction model, the dataset used to train the DL model is based on proteins with deep cavities. Therefore this model may not be suitable for cases where the binding sites are located on flat surfaces. While being a newly released method, Kalasanty has already been subjected to many benchmarks in other method development studies. In addition, relevance of Kalasanty in enzyme engineering has been discussed in this review (Singh et al., Reference Singh, Malik, Gupta and Srivastava2021).
Following the works of DeepSite (Jiménez et al., Reference Jiménez, Doerr, Martínez-Rosell, Rose and De Fabritiis2017), FRSite (Jiang et al., Reference Jiang, Wei, Zhang, Wang, Wang and Li2019), and Kalasanty (Stepniewska-Dziubinska et al., Reference Stepniewska-Dziubinska, Zielenkiewicz and Siedlecki2020), Mylonas et al. (Reference Mylonas, Axenopoulos and Daras2021) developed DeepSurf. DeepSurf uses a variant (Dimou et al., Reference Dimou, Ataloglou, Dimitropoulos, Álvarez and Daras2019) of the DL network architecture known as residual network (ResNet) (He et al., Reference He, Zhang, Ren and Sun2016). The authors propose an 18-layer ResNet to which the surface representation of the target protein is passed in the form of a 3D grid with physiochemical features. The network then predicts a binding site score for the surface presenting points. These points are then clustered and ranked to predict the binding site. The network was trained on a set of 15,182 structures from the sc-PDB database. The distance between the predicted and the real binding site centre and other metrics were used to evaluate DeepSurf. Furthermore, DeepSurf was benchmarked against DL-based methods such as DeepSite, FRSite and Kalasanty as well non-ML methods. The authors showed that compared to DeepSurf, Kalasanty was unable to predict binding sites for a large number of proteins. Interestingly, DeepSite with its shallow neural network layer performed similarly to DeepSurf. While the non-ML methods such as Fpocket (geometry-based) and AutoSite (energy-based) (Ravindranath and Sanner, Reference Ravindranath and Sanner2016) performed poorly, COFACTOR (template-based) (Roy et al., Reference Roy, Yang and Zhang2012) performed similarly when compared to DeepSurf. However, all ML methods generally tend to predict a smaller number of binding sites that are closer to the actual binding sites compared to non-ML methods. DeepSurf has been mentioned in several other studies. In most of these cases, DeepSurf was used as benchmark for other method development and/or other reviews.
Kandel et al. (Reference Kandel, Tayara and Chong2021) developed PUResNet where the protein structure is also treated as a 3D image and the binding site as the object within the image. PUResNet employs variants of two DL architectures, ResNet and U-Net, to address this issue. PUResNet was trained on a set of 5,020 protein structures from the sc-PDB database, and evaluated with DCC, DVO and proportion of ligand inside binding pocket (PLI). The authors also benchmarked their method against Kalasanty, developed by Stepniewska-Dziubinska et al., with a k-fold cross-validation and accuracy test. Briefly, k-fold cross-validation is a method employed to estimate the predictive power of the model on new data. From the benchmarking results, it was observed that PUResNet exhibited slightly better performance during the cross-validation and accuracy test than Kalasanty, despite the fact that PUResNet was trained with only a fraction (1/3) of the dataset used to train Kalasanty. PUResNet has been referenced in several other method development studies as well as other review articles, including a review about pharmacological chaperones for rare diseases (Scafuri et al., Reference Scafuri, Verdino, D’arminio and Marabotti2022).
Along with primary binding site prediction, allosteric site prediction has also been an active area of study (Goncearenco et al., Reference Goncearenco, Mitternacht, Yong, Eisenhaber, Eisenhaber and Berezovsky2013; Panjkovich and Daura, Reference Panjkovich and Daura2014; Greener and Sternberg, Reference Greener and Sternberg2015; Song et al., Reference Song, Liu, Huang, Lu, Shen, Zhang and Zhang2017; Tian et al., Reference Tian, Jiang and Tao2021). The term allosteric binding site refers to a site other than a protein’s primary (orthosteric) active site where a compound can bind, resulting in conformational and dynamic changes (Srinivasan et al., Reference Srinivasan, Forouhar, Shukla, Sampangi, Kulkarni, Abashidze, Seetharaman, Lew, Mao, Acton, Xiao, Everett, Montelione, Tong and Balaram2014). The incorporation of these dynamic changes is critical to accurately predict allosteric sites. The main difference between orthosteric and allosteric ligand binding site prediction algorithms is the inclusion of protein dynamics. Additionally, the algorithms may differ based on the dataset that the DL model was trained on, as the quality and robustness of the data significantly impacts the algorithms’ features and results. Here we review BiteNet (Kozlovskii and Popov, Reference Kozlovskii and Popov2020), a DL-based method for detecting allosteric sites of target proteins from their dynamic ensembles. Similar to other site prediction methods, this work is also inspired by the applications of DL in computer vision (Islam et al., Reference Islam, Rahman, Rahman, Dey and Shoyaib2016). In BiteNet, the input structure of the target protein is placed on a grid and processed with a neural network composed of 10 3D convolutional layers. BiteNet then outputs the centre of the predicted allosteric binding sites, assigns scores and identifies the neighbouring residues within 6 Å radius. BiteNet captures protein dynamics through an ensemble of protein structures gathered from molecular dynamic simulations. When the input is an ensemble of protein structures, BiteNet employs three different clustering algorithms to group the predicted binding site centres and their neighbouring residues and ranks the clusters from most to least probable. The convolutional neural network in BiteNet was trained on 5,946 structures from the PDB. To test for efficiency and accuracy, the authors tested BiteNet’s predictive power on the P2X3 receptor of the ATP-gated cation channel family, the epidermal growth factor receptor of the kinase family, and the adenosine A2A receptor of the G-protein coupled receptor family. In all three cases, it was observed that BiteNet was able to detect conformation-specific allosteric binding sites. Furthermore, BiteNet was benchmarked against FPocket (Le Guilloux et al., Reference Le Guilloux, Schmidtke and Tuffery2009), MetaPocket (Huang, Reference Huang2009), DeepSite (Jiménez et al., Reference Jiménez, Doerr, Martínez-Rosell, Rose and De Fabritiis2017), SiteHound (Hernandez et al., Reference Hernandez, Ghersi and Sanchez2009) and P2Rank (Krivák and Hoksza, Reference Krivák and Hoksza2018) for precision and computational speed on an independent benchmark dataset. In terms of speed and precision, BiteNet outperformed all other methods. BiteNet has been referenced in several other method development studies.
From this summary, it is evident that DL-based methods for binding site prediction have continuously outperformed traditional methods. A summary of the DL-based binding site prediction methods reviewed in this article is presented in Table 2. While these methods are highly successful, they are also limited. For instance, the dataset used to train most of these methods ignores noncanonical amino acids and heavy metals. Thus, these models are still not generalised enough to encompass the astronomical complexities in drug discovery and there is still a huge potential for improvement in the future.
Table 2. Summary of binding site prediction methods
Binding affinity prediction
In SBDD, an accurate prediction of protein-ligand binding affinity can facilitate the discovery of novel drug-like compounds (Fig. 3). In order to achieve such a prediction, a small molecule is typically docked to the target protein (Fig. 3a) with a molecular docking program. Then a score function (SF) is employed to predict the protein-ligand binding affinity (Fig. 3b). Such SFs (if accurate and fast) allow researchers to screen a large number of compounds to identify potential drug-like small molecules. Over the years, several SFs have been developed to approximate the interaction energy between protein and ligand (Jones et al., Reference Jones, Willett, Glen, Leach and Taylor1997; Jain, Reference Jain2003; Friesner et al., Reference Friesner, Banks, Murphy, Halgren, Klicic, Mainz, Repasky, Knoll, Shelley, Perry, Shaw, Francis and Shenkin2004; Shivakumar et al., Reference Shivakumar, Williams, Wu, Damm, Shelley and Sherman2010; Trott and Olson, Reference Trott and Olson2010; Wang et al., Reference Wang, Wu, Deng, Kim, Pierce, Krilov, Lupyan, Robinson, Dahlgren, Greenwood, Romero, Masse, Knight, Steinbrecher, Beuming, Damm, Harder, Sherman, Brewer, Wester, Murcko, Frye, Farid, Lin, Mobley, Jorgensen, Berne, Friesner and Abel2015). These can be classified into physics-based, empirical, and knowledge-based SFs. Physics-based methods include scoring functions based on force fields, solvation models and quantum mechanics (QM) methods. In physics-based SFs, interactions between the protein and the ligand are calculated based on terms such as van der Waals interaction, electrostatic interaction, torsion entropy, solvent effects and others. Due to the limited accuracy of these terms, QM methods hybridised with molecular mechanics (QM-MM) methods have also been utilised to address covalent interactions, polarisation and charge transfer processes (Hayik et al., Reference Hayik, Dunbrack and Merz2010). While QM-MM methods are more accurate, they are also computationally expensive and impractical for screening large number of compounds. Empirical SFs estimate binding affinity by making simplistic approximations to the physics-based energy terms such as hydrogen bonds, hydrophobic effects, steric clashes and other important interactions in a protein-ligand system (Böhm, Reference Böhm1994). These terms are usually associated with weights that are optimised using experimental binding affinity data. Due to the simplicity of the energy terms, empirical score functions are typically less computationally expensive. Knowledge-based SFs are derived based on a statistical analysis of protein-ligand complexes from available 3D structures. In knowledge-based SFs, the frequency of different atom pairs occurring within different distances is assumed to play a key role in the interaction of the atoms. These frequencies are then converted to a distance-dependent potential of mean force (Muegge, Reference Muegge2006). Knowledge-based SFs are also computationally inexpensive and have similar accuracies compared to physics-based methods. However, since knowledge-based SFs do not directly account for experimental binding affinity data, discrepancies between predicted and experimental binding affinity are frequently observed (Stahl and Rarey, Reference Stahl and Rarey2001). Recently, DL-based SFs have also been developed to predict binding affinity and have been shown to outperform traditional SF methods. These models are usually trained on a large dataset with known protein-ligand binding affinity. In DL-based SFs, feautrizing the protein-ligand system plays a key role in its ability to correctly predict the binding affinity. Here, we briefly summarise some of the DL-based methods that have been successful in predicting protein-ligand binding affinity.

Fig. 3. Illustration of computational binding affinity prediction. (a) Small molecule is docked into a target protein. (b) The binding site and the small molecule are then characterised with many features in order to predict the binding affinity. The atoms in the small molecule are shown in grey, blue and red for carbon, nitrogen and oxygen, respectively. The carbon, nitrogen, oxygen and sulphur of the binding site residues are shown in blue, purple, red and yellow, respectively. Bonds in the small molecule ligand are shown in black.
CNNs also serve as popular architectures in binding affinity prediction. Recent examples include variants of traditional 3D-CNNs and ResNet algorithms. Additional CNN-based architectures developed to solve binding affinity prediction are SqueezeNet (Iandola et al., Reference Iandola, Han, Moskewicz, Ashraf, Dally and Keutzer2016) and ShuffleNet (Zhang et al., Reference Zhang, Zhou, Lin and Sun2018). SqueezeNet is a convolution neural network that utilises a design strategy aimed at reducing the number of parameters. Fewer parameters offer several advantages such as: more efficient distributed training, smaller memory storage requirements, and less communication required for over-the-air model updates. SqueezeNet accomplishes its smaller size via Fire modules which replace traditional 3 × 3 filters (kernels) with 1 × 1 filters. ShuffleNet is a CNN architecture that was designed for mobile devices with very limited computing power. In order to reduce computational cost and maintain accuracy, ShuffleNet employs pointwise group convolution and channel shuffle. Grouped convolution makes use of a multiple kernels (filters) per layer, resulting in multiple channel output per layer. This technique led to wider networks helping learn low- and high-level features (Xie et al., Reference Xie, Girshick, Dollár, Tu and He2017), and it has been shown to improve classification accuracy (Xie et al., Reference Xie, Girshick, Dollár, Tu and He2017). Channel shuffle is an operation that helps information flow across feature channels. We review several notable recent examples of these architectures below.
Jiménez et al. (Reference Jiménez, Škalič, Martínez-Rosell and De Fabritiis2018) developed K DEEP, a DL-based method that relies on 3D-convolutional neural network (3D-CNN), to predict protein-ligand binding affinity. The DL architecture in K DEEP is designed based on the image classification DL architecture SqueezeNet (Iandola et al., Reference Iandola, Han, Moskewicz, Ashraf, Dally and Keutzer2016). In K DEEP, residues of the target protein at the binding site and the ligand are placed on a grid. Next, each atom of these interacting residues and the ligand are featurized by eight pharmacophoric-like properties. These descriptors were used to train the K DEEP network on a training dataset that was composed of 3,767 protein-ligand complexes from the PDBbind (Wang et al., Reference Wang, Fang, Lu, Yang and Wang2005) database. Once the network was trained, K DEEP was benchmarked against RF-Score (Ballester and Mitchell, Reference Ballester and Mitchell2010), X-Score (Wang et al., Reference Wang, Lai and Wang2002), and CyScore (Cao and Li, Reference Cao and Li2014) (three other non-DL-based binding affinity prediction methods) for performance and accuracy. Briefly, RF-score is a random forest model trained on a large dataset of molecular interactions. A low RF-score generally corresponds to more favourable interactions. X-Score is an empirical score function based on terms such as van der Waals interactions, hydrogen bonding, deformation penalties, and hydrophobic effects. X-Score values are reported as the Gibbs free energy (ΔG) of binding in the unit of kcal mol−1. Therefore a lower value is indicative of more energetically favourable interactions. Cyscore is another empirical score function that has been optimised based on hydrophobic free energy, van der Waals interaction energy, hydrogen-bond energy and the ligand’s entropy. Cyscore values are comparable to the ΔG (kcal mol−1) of binding. Therefore, a lower Cyscore indicates a higher binding affinity. Compared to other binding affinity prediction methods, K DEEP exhibited the lowest root mean square error (RMSE) with respect to the predicted and experimental affinity. The Pearson’s correlation coefficient of K DEEP was higher than that of X-Score and CyScore but was very similar to that of RF-Score. Furthermore, when these methods were benchmarked against independent datasets that the network had not seen more, K DEEP underperformed compared to RF-Score. Binding affinity predictions from K DEEP are reported as the ΔG (kcal mol−1) of binding. K DEEP is a highly successful program and has been used as a tool in numerous studies. A few examples where K DEEP has been successfully utilised are: to engineer maltooligosaccharide-forming amylases and optimise the biosynthesis of maltooligosaccharides (Ding et al., Reference Ding, Zhao, Han, Li, Gu and Li2022), to determine the inhibitory properties of antiviral drugs and their structural analogues towards coronavirus (Kalamatianos, Reference Kalamatianos2022), during virtual screening (of lead compounds and analogs) to chemically inhibit processes controlled by RecA protein of Acinetobacter baumannii (Tiwari, Reference Tiwari2022) and so forth. While K DEEP is a versatile SF for binding affinity predictions, the authors argued that additional exploration of the network architecture and a thorough featurization of the molecular descriptor could further improve the SF’s performance.
Pafnucy (Stepniewska-Dziubinska et al., Reference Stepniewska-Dziubinska, Zielenkiewicz and Siedlecki2018), developed by Stepniewska-Dzuibinska et. al, is another DL model for predicting binding affinity of protein-ligand complexes. Similar to K DEEP, Pafnucy’s 3D-CNN also treats the ligand and interacting residues of the target protein as a 3D image. This information is then used to create a 3D grid and each atom in the grid is featurized with 19 atomic properties. The network was trained on experimental dissociation constant values of 11,906 protein-ligand complexes (from the PDBbind database) and then evaluated for its performance using RMSE, mean absolute error (MAE), Pearson correlation coefficient, and standard deviation in regression (SD). Therefore, the predicted values from Pafnucy are comparable to experimental dissociation constants. Furthermore, the prediction power of Pafnucy was benchmarked against binding affinity prediction methods such as X-Score, RF-Score, ChemScore, PLP1, ChemPLP (Verdonk et al., Reference Verdonk, Cole, Hartshorn, Murray and Taylor2003), and G-Score (SYBYL 2022). Briefly, ChemScore is an empirical score function based on simple contact terms, a simplistic model for hydrogen bonds, and a penalty function based on flexibility. PLP1 is also an empirical score function that uses terms to account for hydrogen bonds, van der Waals interactions, repulsion potentials, and piecewise linear potentials to model the steric complementarity between protein and ligand. ChemPLP is another empirical score function that is very similar to PLP1. However, in addition to the terms in PLP1, ChemPLP also considers the distance and angle-dependent hydrogen and metal bonding terms from ChemScore. G-Score is a force-field based score function. It is composed of a protein-ligand complexation term (optimised Lenard-Jones potential), a hydrogen bonding term (approximated) and an internal energy term (approximated). All score values from ChemScore, PLP1, ChemPLP, and G-score are reported as the ΔG (kcal mol−1) of binding, therefore lower scores are considered as more favourable interactions. Pafnucy has been successfully used to study the interaction of the mannose receptor of macrophages with carbohydrate ligands (Zlotnikov and Kudryashova, Reference Zlotnikov and Kudryashova2022), as well as has been used as benchmark to develop several newer methods. Pafnucy outperformed all other methods except for RF-Score v3 (Li et al., Reference Li, Leung, Wong and Ballester2015). However, the observed difference in performance was negligible. Additionally, Pafnucy outperformed all 20 binding affinity prediction methods that were used in the CASF-2013 (Li et al., Reference Li, Han, Liu and Wang2014) benchmark. Besides binding affinity prediction, the authors of Pafnucy argue that their model may also be helpful to guide ligand optimization during molecular docking.
Methods that employ DL techniques usually require a large amount of data in order for the method to be properly trained. Thus, limited data could pose a challenge. DeepAtom, a DL method for binding affinity prediction of protein-ligand complexes, developed by Li et al. (Reference Lindert, Li, Sykes and Mccammon2019) addresses this concern with a novel 3D-CNN architecture. The authors of DeepAtom developed an efficient 3D-CNN architecture based on ShuffleNet (Zhang et al., Reference Zhang, Zhou, Lin and Sun2018). Briefly, ShuffleNet is an extremely computation-efficient CNN architecture that is designed for cases with limited computational resources. Using this architecture allowed DeepAtom’s network to be designed with fewer trainable parameters, deep layers, and limited training data. Similar to other methods, DeepAtom also treats the protein-ligand complex as a 3D image. In DeepAtom the interacting residues of the target protein (defined from the centre of the ligand) as well as the ligand are voxelized in a grid. The atoms occupying the voxels are feauturized with 12 atomic properties. To evaluate DeepAtom, the authors first trained DeepAtom, Pafnucy, and RF-Score with 3,390 complexes from the PDBbind database. These models were then evaluated with Pearson correlation coefficient, MAE, RMSE and SD. DeepAtom showed significant improvement over Pafnucy and slight improvement over RF-Score in all metrics. Next, the authors re-trained DeepAtom, Pafnucy and RF-Score on a larger training dataset (9,383 complexes) and evaluated all three models for performance. DeepAtom performed significantly better than both Pafnucy and RF-Score. Since the networks in DeepAtom and Pafnucy are based on 3D-CNN, it was observed that a well-designed network architecture (DeepAtom) can yield better predictions with limited data suggesting that effective training in the deep layers results in improvements in the capacity for learning and generalisation. DeepAtom has been employed in several method development studies (as benchmark) and other review articles.
One key factor of a successful DL scoring function is being able to train the model on features that are important to protein-ligand binding affinity. While the methods discussed so far focused on features that localise to the ligand binding site, OnionNet, developed by Zheng et al. (Reference Zheng, Fan and Mu2019), takes a different approach. The authors treat the protein-ligand complex as a 2D image with one colour channel and use a 2D-CNN architecture to train the binding affinity prediction model. OnionNet is able to capture both local and non-local interactions between the protein and ligand by grouping contacts between each atom of the ligand to atoms in the protein in a set of 60 distance groups. In this way, OnionNet uses 3,840 features to define local and non-local interaction between the target protein and ligand. The network was trained on 11,906 protein-ligand complexes from the PDBbind database. Therefore, predicted values from OnionNet are comparable to experimental dissociation constants. Following the training, the prediction model was evaluated for accuracy and also compared to other ML and non-ML binding affinity prediction methods. In terms of RMSE and Pearson correlation coefficient, OnionNet outperformed Pafnucy and RF-Score. While K DEEP performed slightly better than OnionNet, OnionNet significantly outperformed other non-ML SFs used in the CASF-2013 benchmark. The authors further showed that both the local and non-local interactions contribute significantly to the prediction capability of the model. OnionNet has been utilised as a benchmark during the development of several binding affinity prediction methods.
AK-Score developed by Kwon et al. (Reference Kwon, Shin, Ko and Lee2020) is yet another binding affinity prediction method that was developed based on the ResNet (He et al., Reference He, Zhang, Ren and Sun2016) architecture. Similar to other methods, the protein-ligand system is represented as a 3D image on a grid, with each atom on the grid being defined with eight atomic properties. However, the network architecture uses an ensemble-based approach to perform the binding affinity prediction. AK-Score utilises 20 independently trained networks to make predictions and the average of those values is considered as the final prediction. AK-Scores are reported as the ΔG (kcal mol−1) of binding, therefore a low score corresponds to a more energetically favourable interaction. The networks were trained on 3,772 protein-ligand complexes from the PDBbind database. AK-Score was then evaluated on the CASF-2016 benchmark dataset with Pearson correlation coefficient, Spearman correlation coefficient, Kendall tau, predictive index metrics, MAE, and RSME. AK-Score with ensemble-based networks was benchmarked against AK-Score with a single network and K DEEP. These results showed that the AK-Score ensemble network model had a lower MAE and RMSE as well as higher Pearson correlation coefficient, Spearman correlation coefficient, Kendall tau and predictive index as compared to other methods. Furthermore, from feature analysis, it was shown that the excluded volume of atoms, spatial distribution of hydrophobic and aromatic atoms of the ligand and protein, as well as the distribution of hydrogen bond acceptors of the protein were particularly important for the network to accurately determine the binding affinity. Therefore, more accurate calculations of such descriptors may play an important role in improving the ensemble-based network of AK-Score. AK-Score has been reviewed in several reviews and its prediction quality has been compared in other method development studies.
Similar to AK-Score, RosENet developed by Hassan-Harrirou et al. (Reference Hassan-Harrirou, Zhang and Lemmin2020), also uses an ensemble-based 3D-CNN. However, RosENet uses a hybrid approach where it employs the 3D-CNN to combine molecular mechanics energies calculated from the Rosetta force field (Alford et al., Reference Alford, Leaver-Fay, Jeliazkov, O’Meara, Dimaio, Park, Shapovalov, Renfrew, Mulligan, Kappel, Labonte, Pacella, Bonneau, Bradley, Dunbrack, Das, Baker, Kuhlman, Kortemme and Gray2017) with molecular descriptors. Briefly, the Rosetta software suite includes algorithms for computational modelling and analysis of protein structures and has enabled scientific advances in areas such as protein design, enzyme design, molecular docking and structure prediction (Leman et al., Reference Leman, Weitzner, Lewis, Adolf-Bryfogle, Alam, Alford, Aprahamian, Baker, Barlow, Barth, Basanta, Bender, Blacklock, Bonet, Boyken, Bradley, Bystroff, Conway, Cooper, Correia, Coventry, Das, De Jong, Dimaio, Dsilva, Dunbrack, Ford, Frenz, Fu, Geniesse, Goldschmidt, Gowthaman, Gray, Gront, Guffy, Horowitz, Huang, Huber, Jacobs, Jeliazkov, Johnson, Kappel, Karanicolas, Khakzad, Khar, Khare, Khatib, Khramushin, King, Kleffner, Koepnick, Kortemme, Kuenze, Kuhlman, Kuroda, Labonte, Lai, Lapidoth, Leaver-Fay, Lindert, Linsky, London, Lubin, Lyskov, Maguire, Malmström, Marcos, Marcu, Marze, Meiler, Moretti, Mulligan, Nerli, Norn, Ó’Conchúir, Ollikainen, Ovchinnikov, Pacella, Pan, Park, Pavlovicz, Pethe, Pierce, Pilla, Raveh, Renfrew, Burman, Rubenstein, Sauer, Scheck, Schief, Schueler-Furman, Sedan, Sevy, Sgourakis, Shi, Siegel, Silva, Smith, Song, Stein, Szegedy, Teets, Thyme, Wang, Watkins, Zimmerman and Bonneau2020). The network architecture of RosENet was developed based on the ResNet architecture. Similar to previous methods, protein-ligand complexes were represented in a 3D grid. Atoms of the protein-ligand complex were featurized with 12 molecular descriptors from HTMD (Doerr et al., Reference Doerr, Harvey, Noé and De Fabritiis2016) and 6 molecular energies obtained from Rosetta. The network was then trained on a curated dataset of 4,463 protein-ligand complexes obtained from the PDBbind database. Therefore, the predicted values from RosENet are comparable to experimental dissociation constants. To evaluate RosENet with respect to the quality of its predictions, the network was first optimised based on a combination of multiple features and architectures, and then compared to Pafnucy and OnionNet. The authors tested two neural network architectures: ResNet (a deep residual neural network) and SqueezeNet (a smaller network architecture). RosENet showed a lower RMSE with the ResNet architecture than it did with the SqueezeNet architecture. Thus, the authors argue that the network implemented in the SqueezeNet architecture was not deep enough to generalise the molecular mechanics energies that were used to featurize the complex. RosENet was also trained with a smaller number of features, leading to a decrease in the prediction quality, however, this change was considered to be insignificant. Overall, RosENet performed better than Pafnucy and comparably to OnionNet. This suggested that the molecular energies described in RosENet may be able to represent similar local and non-local interactions described by the OnionNet. Finally, the RMSE of RosENet decreased when the average of the five best predictions was used. In future studies, the authors of RosENet propose to improve their molecular features by extracting additional dynamic properties from molecular dynamics simulations. RosENet has been featured in other method development studies and as well as review articles.
Accurate prediction of binding affinity is a key step during the drug discovery process. Despite several limitations, recently developed DL-based methods for binding affinity prediction have generally significantly outperformed traditional methods. A summary of such DL-based methods as reviewed in this article can be found in Table 3. These studies indicate the availability of successful in silico tools for rapid and accurate prediction of protein-ligand binding affinity. Additionally, a few ML-based methods (Aggarwal and Koes, Reference Aggarwal and Koes2020; Bao et al., Reference Bao, He and Zhang2021) have also been trained to predict ‘the root mean square deviation (RMSD)’ of a docked structure with reference to the native bound structure. Deep Binding Structure RMSD Prediction model (DeepBSP) (Bao et al., Reference Bao, He and Zhang2021) is one such method that employs a 3D-CNN (same model architecture as K DEEP) to achieve this purpose. DeepBSP exhibited accuracy on predicting RMSD. Furthermore, the docking power of DeepBSP (through model selection with lowest predicted RMSD), outperformed all other scoring functions benchmarked in CASF-2016. While these methods have been successful, this field continues to grow and evolve with new advancements in ML.
Table 3. Summary of binding affinity prediction methods

Conclusion
Over recent years, ML has become an increasingly popular field of study. Application of these methods to biological problems will only continue to grow as academic and industry users create better tools to predict biomolecular structure, treat disease, and improve public health. In this work, we summarised recent DL applications in three structure-based drug discovery categories: de novo drug design, binding site prediction and binding affinity prediction. In the interest of the reader, we also provide links to the various methods’ publicly available source code or webservers. The use of ML (more recently DL) methods is not limited solely to SBDD, and they have also been applied to all other areas of the drug discovery pipeline such as LBDD (Bahi and Batouche, Reference Bahi and Batouche2018), lead optimization (de Souza Neto et al., Reference De Souza Neto, Moreira-Filho, Neves, Maidana, Guimarães, Furnham, Andrade and Silva2020), and assessment of absorption (Shin et al., Reference Shin, Jang, Nam, Lee and Lee2018), metabolism (Wang et al., Reference Wang, Liu, Shen, Jiang, Wang, Li and Li2020; Litsa et al., Reference Litsa, Das and Kavraki2021), binding kinetics (De Benedetti and Fanelli, Reference De Benedetti and Fanelli2018; Mardt et al., Reference Mylonas, Axenopoulos and Daras2018; Gao et al., Reference Gao, Jiang, Fu and He2019; Nunes-Alves et al., Reference Nunes-Alves, Kokh and Wade2020; Feizpour et al., Reference Feizpour, Marstrand, Bastholm, Eirefelt and Evans2021; Obeid et al., Reference Obeid, Madžarević, Krkobabić and Ibrić2021; Hoffmann et al., Reference Hoffmann, Gerst, Cseresnyés, Foo, Sommerfeld, Press, Bauer and Figge2022), efficacy (Lin et al., Reference Lin, Kuo, Liu, Yu, Yang and Tsai2018; Benning et al., Reference Benning, Peintner, Finkenzeller and Peintner2020; Li et al., Reference Li, Wu, Zhao, Zhang, Shao, Wang, Qiu and Li2021a; Zhu et al., Reference Zhu, Wang, Wang, Gao, Guo, Gao, Liu, Yu, Wang, Kong, An, Liu, Sun, Huang, Zhou, Zhang, Zheng and Xie2021) and toxicity properties (Ferreira and Andricopulo, Reference Ferreira and Andricopulo2019; Liu et al., Reference Liu, Sun, Jia, Ma, Xing, Wu, Gao, Sun, Boulnois and Fan2019; Shi et al., Reference Shi, Yang, Huang, Chen, Kuang, Heng and Mei2019; Cáceres et al., Reference Cáceres, Tudor and Cheng2020; Feinberg et al., Reference Feinberg, Joshi, Pande and Cheng2020). While these studies are not covered in this review, interested readers are encouraged to review these recent works as well.
With the expansion of large datasets, DL algorithms will only become more accurate, thereby increasing cost-effectiveness and time efficiency. However, a major critique of DL methods is their inherit ‘black-box’ nature. In order for the use of large datasets to be truly successful, we must begin to understand the nature of DL algorithms and the connections they make. Another common pitfall of any ML algorithm is the quality of its training dataset(s). In any biological application, an algorithm may easily become hyper-specific to a certain motif if it is not exposed to an unbiased training set. This lack of generalisation prevents the algorithm from solving real-world problems.
Recently the company DeepMind and the Baker research group have released AlphaFold (Jumper et al., Reference Jumper, Evans, Pritzel, Green, Figurnov, Ronneberger, Tunyasuvunakool, Bates, Žídek, Potapenko, Bridgland, Meyer, Kohl, Ballard, Cowie, Romera-Paredes, Nikolov, Jain, Adler, Back, Petersen, Reiman, Clancy, Zielinski, Steinegger, Pacholska, Berghammer, Bodenstein, Silver, Vinyals, Senior, Kavukcuoglu, Kohli and Hassabis2021) and RoseTTAFold (Baek et al., Reference Baek, Dimaio, Anishchenko, Dauparas, Ovchinnikov, Lee, Wang, Cong, Kinch, Schaeffer, Millán, Park, Adams, Glassman, Degiovanni, Pereira, Rodrigues, Dijk, Ebrecht, Opperman, Sagmeister, Buhlheller, Pavkov-Keller, Rathinaswamy, Dalwadi, Yip, Burke, Garcia, Grishin, Adams, Read and Baker2021), DL algorithms for protein structure prediction. These algorithms have been credited with ‘solving the protein folding problem’. Given the impressive prediction capabilities of these tools, it is natural to investigate how relevant such DL-based protein conformations are for drug design. For example, recently the crystal structure of the σ2 receptor has been reported (Alon et al., Reference Alon, Lyu, Braz, Tummino, Craik, O’Meara, Webb, Radchenko, Moroz, Huang, Liu, Roth, Irwin, Basbaum, Shoichet and Kruse2021). In this study, the authors also screened a large library of compounds to the X-ray structure (through molecular docking), which resulted in the identification of around 130 active compounds. However, the molecular docking of these same hits scored relatively poorly against the AlphaFold model. This suggests that additional efforts are needed to employ predicted structures successfully in drug discovery. Within the last year, the CEO of DeepMind announced the plan of applying their knowledge of artificial intelligence to drug discovery with the formation of a new company called Isomorphic Labs (https://www.isomorphiclabs.com). This year another company, CHARM Therapeutics (https://charmtx.com), in the UK have launched DragonFold. DragonFold is the first DL algorithm to rapidly predict protein-ligand co-folding. While we wait on more details of these developments, they however foreshadow a very promising future for the field of computer-aided drug discovery, as the brightest minds from industry and academia continue to collaborate and improve upon ML applications. While the future of the field appears very promising, we must also keep in mind the ethics of good science. As demonstrated by Urbina et al. (Reference Urbina, Lentzos, Invernizzi and Ekins2022), artificial intelligence was utilised to predict the toxicity of small molecules. The algorithm developed by the Collaborations team predicted many compounds with higher toxicity as compared to the nerve agent VX. The team calls for a strengthening of ethical training of students, as well as for artificial intelligence drug discovery companies to create a code of conduct to properly train employees and establish protection measures for their technology.
Open Peer Review
To view the open peer review materials for this article, please visit http://doi.org/10.1017/qrd.2022.12.










Comments
Comments to Author: Lindert and coworkers presented a thorough review of recent deep learning trends in computer-aided drug design with focus on de novo drug design, binding site prediction and binding affinity prediction of small molecules.
It can be potentially improved regarding the following:
1. It would help to explain briefly various scores, including the Q-value, SA and QED scores, Vinardo score, RF-score, X-score, cyScore, Chem score, AK-Score, etc.
How are they defined, what are the ranges and what values are needed for "good" predictions?
2. What are the main difference(s) in the deep learning algorithms for predicting allosteric sites compared with predicting orthosteric/primary ligand binding sites.
3. Although drug binding kinetics and efficacy are not covered in the review, it would help to still briefly comment on deep learning studies of these and related drug design aspects.