Introduction
Tritrichomonas foetus is an important bovine venereal parasite which causes reproductive issues including spontaneous abortion, pyometra and infertility (Dąbrowska et al., Reference Dąbrowska, Karamon, Kochanowski, Sroka, Zdybel and Cencek2019), imposing a significant economic burden on the beef and dairy industry (Yule et al., Reference Yule, Skirrow and Bonduran1989). Tritrichomonas foetus is also a prevalent cause of diarrhoea in cats (Asisi et al., Reference Asisi, Steiner, Pfister and Kohn2008), and is considered to be synonymous with the porcine gut-associated Tritrichomonas suis (Slapeta et al., Reference Slapeta, Müller, Stack, Walker, Lew-Tabor, Tachezy and Frey2012). Tritrichomonas foetus is a protozoan parasite of the class Tritrichomonadea, which together with Trichomonadea belong to the informal taxonomic group the trichomonads, within the phylum parabasalia (Cepicka et al., Reference Cepicka, Hampl and Kulda2010; Noda et al., Reference Noda, Mantini, Meloni, Inoue, Kitade, Viscogliosi and Ohkuma2012). Tetratrichomonas is a diverse genus within Trichomonadea, which most commonly form symbiotic relationships within the gastrointestinal tract (GIT) of animals including invertebrates, fish, mammals, reptiles, amphibians and birds (Cepicka et al., Reference Cepicka, Hampl, Kulda and Flegr2006). The genus was originally defined morphologically with characteristics including an ovoid cell body and four anterior flagella of unequal length (Honigberg, Reference Honigberg1963). There have been conflicting reports regarding the monophyly of Tetratrichomonas, with some molecular phylogenies placing Trichomonas, Pentatrichomonas and Trichomonoides as ingroups (Cepicka et al., Reference Cepicka, Hampl, Kulda and Flegr2006).
Trichomonads appear to undergo frequent cross-species transmission and transfer to alternative body sites; Pentatrichomonas hominis has been isolated from a wide range of mammals including humans, primates, cats, dogs and bovids (Li et al., Reference Li, Ying, Gong, Li, Yang, Li and Zhang2016; Bastos et al., Reference Bastos, Brener, de Figueiredo, Leles and Mendes-de-Almeida2018; Li et al., Reference Li, Huang, Fang, Ren, Tang, Kan, Liu and Gu2020), and it is thought that the human pathogens Trichomonas tenax and Trichomonas vaginalis arose from cross-species transmission of independent lineages of avian oral parasites to the human mouth and urogenital tract (UGT), respectively (Maritz et al., Reference Maritz, Land, Carlton and Hirt2014; Peters et al., Reference Peters, Das and Raidal2020). Some reports have suggested a degree of fidelity between Tetratrichomonas and host lineages. For example, Cepicka et al. (Reference Cepicka, Hampl, Kulda and Flegr2006) identified 16 Tetratrichomonas lineages of which the majority were fairly limited in the taxonomic range of host species. However, Tetratrichomonas spp. are often not limited to a single host, for example, Tetratrichomonas gallinarum has been identified as a pathological agent in a wide range of bird species (Cepicka et al., Reference Cepicka, Kutigova, Tachezy, Kulda and Flegr2005). Transmission across wider taxonomic ranges are also not unknown, and reports of Tetratrichomonas spp. in the lungs of immunocompetent patients with pulmonary disease (Lin et al., Reference Lin, Ying, Lai, Li, Xue, Zhou and Hu2019) illustrates the potential importance of Tetratrichomonas zoonosis for human health.
There have been several reports of non-Tt. foetus trichomonads isolated from the bull preputial cavity. Morphological and phylogenetic methods have identified these as P hominis, a Pseudotrichomonas sp. and Tetratrichomonas spp., some of which may correspond to the previously described species Tetratrichomonas buttreyi (Dufernez et al., Reference Dufernez, Walker, Noël, Caby, Mantini, Delgado-Viscogliosi, Ohkuma, Kudo, Capron, Pierce, Villanueva and Viscogliosi2007). The most common method for Tt. foetus diagnosis is in vitro culture of preputial washings, and subsequent examination of cultures by light microscopy (Parker et al., Reference Parker, Campbell, McIntosh and Gajadhar2003), and so false positives resulting from non-Tt. foetus trichomonads may be an issue (Dufernez et al., Reference Dufernez, Walker, Noël, Caby, Mantini, Delgado-Viscogliosi, Ohkuma, Kudo, Capron, Pierce, Villanueva and Viscogliosi2007).
This study provides an additional report of a Tetratrichomonas sp. isolated from bull preputial washings in the UK, and reviews previous reports on trichomonad species in the bovine UGT.
Materials and methods
Source of isolates
Samples were collected from bulls in the UK during routine screening for parasites, in accordance with the principles defined in the European Convention for the Protection of Vertebrate Animals used for Experimental and Other Scientific Purposes. To collect samples, the preputial cavity was washed with 30 mL pre-warmed phosphate buffered saline (PBS) (pH 7.2). Washings were pelleted by centrifugation at 300 × g for 10 min, suspended in 5 mL PBS and examined for motile protozoa. Parasites were cultured by inoculating 1 mL of the PBS suspension into 10 mL Clausens medium [20 g L−1 Neopeptone, 10 g L−1 Lab Lemco (Oxoid), 5 g L−1 neutralized liver digest (Oxoid) and 20 g L−1 glucose, pH 7.4], supplemented with 200 units mL−1 penicillin, 200 μg mL−1 streptomycin and 1000 units mL−1 polymixin B, overlaid on a solid medium slope prepared by heating 7 mL horse serum at 75°C for 2 h. Cultures were incubated at 37°C for 7 days, and were examined microscopically on days 4 and 7 for motile protozoa. Positive cultures were passaged in fresh media, and 10 mL culture was harvested by centrifugation at 300 × g for 10 min, fixed in 15 mL 100% ethanol and stored at −20°C. No cryopreserved stock of the isolate was generated.
Molecular sequence typing
Genomic DNA was extracted from ethanol-fixed parasite isolates using the DNeasy ultraclean microbial kit (Qiagen) according to the manufacturer's instructions. Loci for molecular sequence typing were amplified by polymerase chain reaction (PCR) using generic Taq polymerase (NEB). A region containing the 5.8S ribosomal RNA (rRNA) and flanking internal transcribed spacers (ITS) 1 and 2 was amplified using the trichomonad-specific TFR1 and TFR2 primers, and the 18S rRNA gene was amplified using the generic eukaryotic primers Euk 1700 and Euk 1900. Resulting products were cloned into pCR4TOPO using the TOPO TA cloning kit (ThermoFisher Scientific) according to the manufacturer's instructions. Sequences were generated for the inserts from five independent clones by Sanger sequencing (Eurofins) on both strands using the T7 and T3 promoter primers, and additional internal sequencing primers were designed to cover the full length of the 18S rRNA gene for both strands. All primer sequences are listed in Supplementary Table S1.
Phylogenetic analysis
A reference collection of ITS1-5.8S rRNA-ITS2 and 18S rRNA trichomonad sequences were selected from the NCBI RefSeq database (O'Leary et al., Reference O'Leary, Wright, Brister, Ciufo, Haddad, McVeigh, Rajput, Robbertse, Smith-White, Ako-Adjei, Astashyn, Badretdin, Bao, Blinkova, Brover, Chetvernin, Choi, Cox, Ermolaeva, Farrell, Goldfarb, Gupta, Haft, Hatcher, Hlavina, Joardar, Kodali, Li, Maglott, Masterson, McGarvey, Murphy, O'Neill, Pujar, Rangwala, Rausch, Riddick, Schoch, Shkeda, Storz, Sun, Thibaud-Nissen, Tolstoy, Tully, Vatsan, Wallin, Webb, Wu, Landrum, Kimchi, Tatusova, DiCuccio, Kitts, Murphy and Pruitt2016) by BLASTn search (Altschul et al., Reference Altschul, Gish, Miller, Myers and Lipman1990) using the new sequences as a query, and from the literature. Sequences were aligned using Tcoffee (Notredame et al., Reference Notredame, Higgins and Heringa2000), gaps and poorly aligned regions were trimmed automatically using trimAl (Capella-Gutiérrez et al., Reference Capella-Gutiérrez, Silla-Martínez and Gabaldón2009) and resulting alignments were visually inspected for accuracy using Seaview (Gouy et al., Reference Gouy, Guindon and Gascuel2010). Iqtree (Nguyen et al., Reference Nguyen, Schmidt, von Haeseler and Minh2015; Kalyaanamoorthy et al., Reference Kalyaanamoorthy, Minh, Wong, von Haeseler and Jermiin2017) was used to construct maximum likelihood (ML) phylogenies using automatic model selection, and reliability was assessed with 1000 bootstrap replicates. Figures were generated using iTol (Letunic and Bork, Reference Letunic and Bork2019). Alignments used to generate phylogenies are available in the Supplementary data (S1, S2 and S3).
Metatranscriptomics data analysis
Sequence data for total RNA metatranscriptomics sequencing of the cattle rumen was downloaded from NCBI's SRA database (Leinonen et al., Reference Leinonen, Sugawara, Shumway and Collaboration2011), selecting a subset of 14 out of a total of 48 available samples for analysis (accessions SRX5208721–SRX5208732 and SRX5229555). SortMeRNA (Kopylova et al., Reference Kopylova, Noé and Touzet2012) was used to align the reads to the default reference rRNA database, and aligned reads were searched for parasite sequences by BLASTn (Altschul et al., Reference Altschul, Gish, Miller, Myers and Lipman1990), collecting only the top hit for each query.
Results
Between 2014 and 2021, preputial samples from 9637 bulls were screened for trichomonads. For only a single sample, from a 1-year-old bull in South West England, motile, trichomonad-like protozoa were isolated. Morphological examination by light microscopy revealed four anterior flagella. Rearing conditions of the bull are likely to have allowed sexual contact with other bulls and cows.
In order to identify the isolate, sequences were generated for five clones for the 18S rRNA and ITS1-5.8S rRNA-ITS2 loci. For the 18S rRNA locus, three unique clonal sequences were generated which shared 99.5–99.7% identity, suggesting a single-species infection. ML analysis of the 18S rRNA locus placed all clone sequences together within Tetratrichomonas with strong bootstrap support (Supplementary Fig. S1).
For the ITS1-5.8S rRNA-ITS2 locus, there were three unique sequences which shared 95.2–99% identity, suggesting some diversity amongst the parasites present. In agreement with the 18S rRNA locus, ML analysis also placed all the isolates together within Tetratrichomonas with moderate bootstrap support (Supplementary Fig. S2). The isolates were also placed in a separate lineage from urogenital Tetratrichomonas isolates from cattle originating from a previous study (accession AF342742).
To resolve the Tetratrichomonas lineage of the isolates in more detail, ML analysis was performed on the 18S rRNA sequence from a greater taxonomic sampling of Tetratrichomonas spp., including several bull urogenital isolates from previous studies (Fig. 1). The sequences for all clones grouped within Tetratrichomonas lineage 4 as defined by Cepicka et al. (Reference Cepicka, Hampl, Kulda and Flegr2006), which includes one group of bull urogenital isolates previously reported (accession DQ412633).
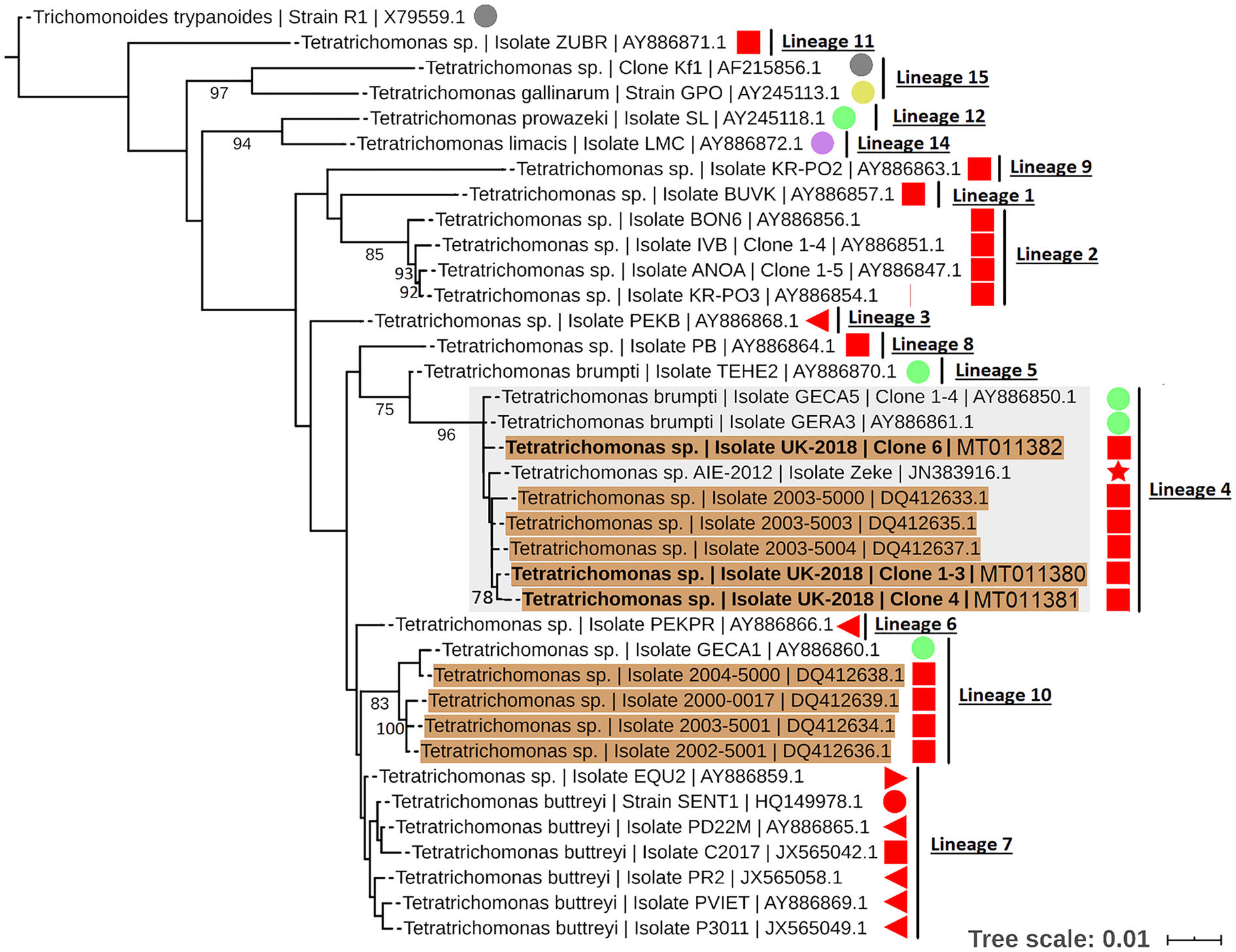
Fig. 1. ML phylogeny (GTR model with empirical base frequencies, invariable sites and discrete gamma model) for Tetratrichomonas spp. based on the 18S rRNA locus, rooted using Trichomitus batrachorum as an outgroup. Bootstrap values (1000 replicates) >70% are shown on branches. New sequences generated in this study are highlighted in bold. Tetratrichomonas lineages as defined by Cepicka et al. (Reference Cepicka, Hampl, Kulda and Flegr2006) are annotated, and the lineage of interest is highlighted. Urogenital isolates are highlighted in orange, and host species are annotated with shapes at the tip labels; insect (grey), mollusc (purple), bird (yellow), reptile (green) and mammal (red). Mammals are subdivided into families; bovine (square), porcine (left triangle), equine (right triangle), primate (circle) and anteater (star). Units for tree scale are inferred substitutions per base pair. Genbank (Benson et al., Reference Benson, Clark, Karsch-Mizrachi, Lipman, Ostell and Sayers2015) accessions for each sequence are shown at the end of tip labels.
In order to investigate the potential gastrointestinal origin of urogenital Tetratrichomonas spp. in cattle, we searched published metatranscriptomics data from the cattle rumen for sequences similar to two hypervariable regions (V4 and V8) amongst Tetratrichomonas 18S rRNA sequences. For the published urogenital Tetratrichomonas isolate 2004–5000 (Dufernez et al., Reference Dufernez, Walker, Noël, Caby, Mantini, Delgado-Viscogliosi, Ohkuma, Kudo, Capron, Pierce, Villanueva and Viscogliosi2007; accession DQ412638) we identified sequences identical to the V8 region, and virtually identical (2 base pair mismatch) to the V4 region. BLAST search results were identical in terms of mismatches, gaps and query coverage for 11 out of 14 samples tested. In contrast, we did not identify any sequences of a similarly highly sequence identity to the urogenital Tetratrichomonas sequence generated in this study. Representative BLAST results for a single sample are shown in Supplementary Table S2, and an alignment between metatranscriptome sequences and urogenital Tetratrichomonas 18S rRNA sequences are shown in Supplementary file S4.
In addition to the isolates identified in this study, there have been numerous reports of trichomonads isolated from the UGT of cattle, summarized in Table 1. A total of 151 trichomonad isolates have been reported. With the exception of isolates obtained from this study, all previous isolates originate from the Americas. Various morphological and molecular techniques have assigned the isolates as Tetratrichomonas and Pentatrichomonas spp., which appear to have been isolated in roughly equal proportions. In contrast, there has only been a single report of a Pseudotrichomonas isolate.
Table 1. Summary of reports of non-Tritrichomonas foetus trichomonads isolated from the cattle UGT
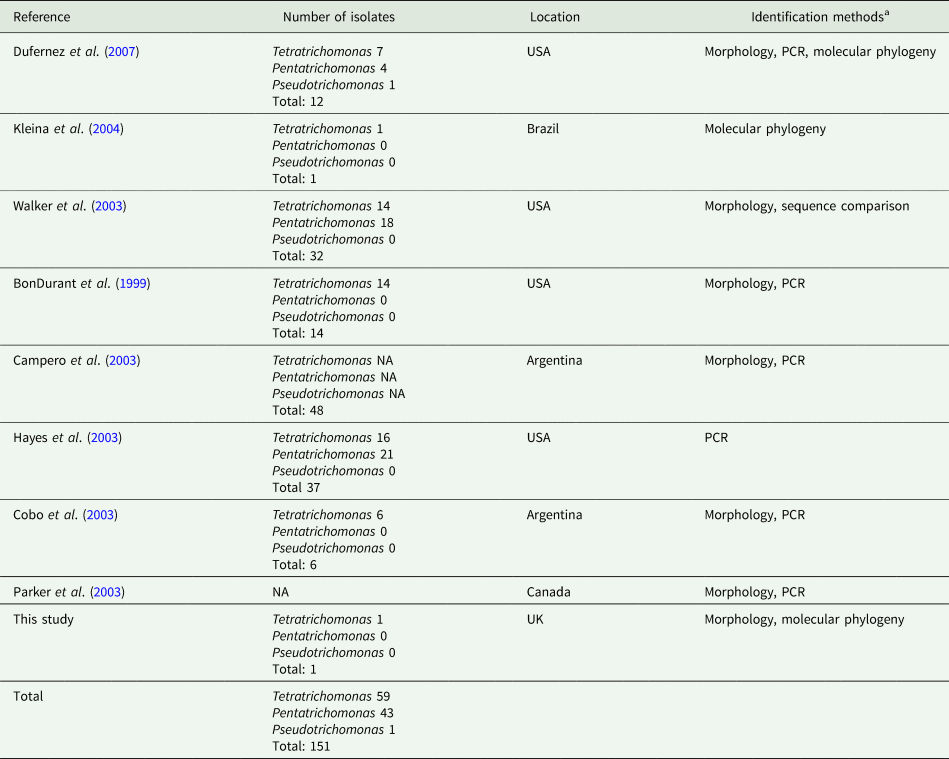
aPCR and phylogenetic data are based on amplification of the ITS1-5.8S rRNA-ITS2 locus, except for Dufernez et al. (Reference Dufernez, Walker, Noël, Caby, Mantini, Delgado-Viscogliosi, Ohkuma, Kudo, Capron, Pierce, Villanueva and Viscogliosi2007) and this study, which also examined the 18S rRNA locus.
Discussion
This study reports the first case of a non-Tt. foetus trichomonad isolated from the bovine UGT in Europe. In agreement with previous reports of cattle urogenital trichomonads (Walker et al., Reference Walker, Hayes, Sawyer, Nordhausen, Van Hoosear and BonDurant2003; Kleina et al., Reference Kleina, Bettim-Bandinelli, Bonatto, Benchimol and Bogo2004; Dufernez et al., Reference Dufernez, Walker, Noël, Caby, Mantini, Delgado-Viscogliosi, Ohkuma, Kudo, Capron, Pierce, Villanueva and Viscogliosi2007), the morphological and sequence data indicate that this isolate is a Tetratrichomonas sp. The phylogenies presented here are largely in agreement with previous studies regarding the interrelationship between trichomonads and the reported lineages of Tetratrichomonas spp. (Cepicka et al., Reference Cepicka, Hampl, Kulda and Flegr2006; Dufernez et al., Reference Dufernez, Walker, Noël, Caby, Mantini, Delgado-Viscogliosi, Ohkuma, Kudo, Capron, Pierce, Villanueva and Viscogliosi2007). Similarly to previous reports (Cepicka et al., Reference Cepicka, Hampl, Kulda and Flegr2006, Reference Cepicka, Hampl and Kulda2010), there is discrepancy between phylogenies based on the 18S rRNA and ITS1-5.8S rRNA-ITS2 in terms of the monophyly of the Tetratrichomonas genus.
Our results support the placement of the new isolates in Tetratrichomonas lineage 4 as defined by Cepicka et al. (Reference Cepicka, Hampl, Kulda and Flegr2006), which corresponds to group B of the bull urogenital Tetratrichomonas isolates defined by Dufernez et al. (Reference Dufernez, Walker, Noël, Caby, Mantini, Delgado-Viscogliosi, Ohkuma, Kudo, Capron, Pierce, Villanueva and Viscogliosi2007). The new isolates were distinct from bull urogenital group A (Dufernez et al., Reference Dufernez, Walker, Noël, Caby, Mantini, Delgado-Viscogliosi, Ohkuma, Kudo, Capron, Pierce, Villanueva and Viscogliosi2007), corresponding to Tetratrichomonas lineage 10 (Cepicka et al., Reference Cepicka, Hampl, Kulda and Flegr2006) and also possibly the bull urogenital isolates described by Walker et al. (Reference Walker, Hayes, Sawyer, Nordhausen, Van Hoosear and BonDurant2003), although this is not well resolved. In addition, the ITS1-5.8S rRNA-ITS2 data suggest some genetic diversity amongst the clonal sequences from the new isolate, as the three clones did not cluster together, potentially indicating a non-clonal infection or heterogeneity amongst multi-copy rRNA genes (Torres-Machorro et al., Reference Torres-Machorro, Hernández, Cevallos and López-Villaseñor2010). These observations overall suggest a degree of genetic heterogeneity amongst cattle urogenital Tetratrichomonas isolates.
Tetratrichomonas lineage 4 was described as mostly limited to the tortoise GIT (Cepicka et al., Reference Cepicka, Hampl, Kulda and Flegr2006). This lineage has also been found in mammals, but excluding the cattle isolates, all others including the elephant isolate SLON (Cepicka et al., Reference Cepicka, Hampl, Kulda and Flegr2006) and the anteater isolate Zeke (Ibañez-Escribano et al., Reference Ibañez-Escribano, Nogal-Ruiz, Delclaux, Martinez-Nevado and Ponce-Gordo2013), originate from zoo animals which may suggest an artificial context for transmission. Isolates from Tetratrichomonas lineage 10 (Cepicka et al., Reference Cepicka, Hampl, Kulda and Flegr2006) have also been found in the bull UGT, and based on morphological data it has been suggested that this corresponds to the previously described cattle gastrointestinal species Te. buttreyi (Dufernez et al., Reference Dufernez, Walker, Noël, Caby, Mantini, Delgado-Viscogliosi, Ohkuma, Kudo, Capron, Pierce, Villanueva and Viscogliosi2007). Intriguingly, Tetratrichomonas lineage 10 also appears to be mostly restricted to tortoises (Cepicka et al., Reference Cepicka, Hampl, Kulda and Flegr2006).
At least two Tetratrichomonas lineages, P. hominis, and a Pseudotrichomonas sp. have been isolated from the cattle UGT (Walker et al., Reference Walker, Hayes, Sawyer, Nordhausen, Van Hoosear and BonDurant2003; Dufernez et al., Reference Dufernez, Walker, Noël, Caby, Mantini, Delgado-Viscogliosi, Ohkuma, Kudo, Capron, Pierce, Villanueva and Viscogliosi2007). These trichomonad isolates alternatively represent either emerging or established urogenital species or sporadic spillover of microbes from an alternative reservoir, most likely the bovine GIT. The possibility of cross contamination during sample collection should also not be discounted for some isolates. The genetic heterogeneity amongst cattle urogenital isolates may support the spillover hypothesis. This is in contrast to parasite species thought to have emerged through cross-species or cross-mucosa transmission, such as Tt. foetus, which shows a remarkable degree of genetic homogeneity suggestive of a recent founder event (Kleina et al., Reference Kleina, Bettim-Bandinelli, Bonatto, Benchimol and Bogo2004). Cattle urogenital trichomonad isolation shows a sporadic geographic pattern (Campero et al., Reference Campero, Rodriguez Dubra, Bolondi, Cacciato, Cobo, Perez, Odeon, Cipolla and BonDurant2003; Kleina et al., Reference Kleina, Bettim-Bandinelli, Bonatto, Benchimol and Bogo2004; Dufernez et al., Reference Dufernez, Walker, Noël, Caby, Mantini, Delgado-Viscogliosi, Ohkuma, Kudo, Capron, Pierce, Villanueva and Viscogliosi2007) which also supports the spillover hypothesis, as the events appear to be unrelated to one another. We identified sequences highly similar to cattle urogenital Tetratrichomonas 18S rRNA (Dufernez et al., Reference Dufernez, Walker, Noël, Caby, Mantini, Delgado-Viscogliosi, Ohkuma, Kudo, Capron, Pierce, Villanueva and Viscogliosi2007) frequently occurring in the cattle rumen metatranscriptome (Li et al., Reference Li, Hitch, Chen, Creevey and Guan2019), providing strong evidence that these urogenital isolates are of gastrointestinal origin. The infrequent urogenital detection of non-Tt. foetus trichomonads in a backdrop of frequent Tt. foetus monitoring (Yao, Reference Yao2013) suggests that the events are rare, although misidentification of other trichomonads as Tt. foetus is also possible.
There are plausible sources for the isolated urogenital trichomonads from the bovine GIT; Tetratrichomonas spp. (Zhang et al., Reference Zhang, Li, Chen, Wu, Meng and Guan2020), such as Te. buttreyi (Dufernez et al., Reference Dufernez, Walker, Noël, Caby, Mantini, Delgado-Viscogliosi, Ohkuma, Kudo, Capron, Pierce, Villanueva and Viscogliosi2007), are known to inhabit the cattle gut, and P. hominis is known to occupy the GIT of a very wide range of animals (Li et al., Reference Li, Ying, Gong, Li, Yang, Li and Zhang2016), including cattle (Li et al., Reference Li, Huang, Fang, Ren, Tang, Kan, Liu and Gu2020). The isolate related to the putatively free-living Pseudotrichomonas keilini (Dufernez et al., Reference Dufernez, Walker, Noël, Caby, Mantini, Delgado-Viscogliosi, Ohkuma, Kudo, Capron, Pierce, Villanueva and Viscogliosi2007) at most represents a very rare event, as only a single isolate has been reported, and so contamination cannot be ruled out. Alternatively, Monoceromonas ruminantium is also relatively closely related (Dufernez et al., Reference Dufernez, Walker, Noël, Caby, Mantini, Delgado-Viscogliosi, Ohkuma, Kudo, Capron, Pierce, Villanueva and Viscogliosi2007) and has been isolated from the GIT of cattle (Hampl et al., Reference Hampl, Cepicka, Flegr, Tachezy and Kulda2004), suggesting that the lineage could be host-associated and that P. keilini is an exception or mislabelled as free-living. The presence of gastrointestinal-associated bacteria at the same site as urogenital trichomonad isolation provides further evidence for their gut origin (Cobo et al., Reference Cobo, Campero, Mariante and Benchimol2003). A route of gastrointestinal to urogenital transmission through sexual mounting behaviour, potentially between young bulls, has been suggested (BonDurant et al., Reference BonDurant, Gajadhar, Campero, Johnson, Lun, Nordhausen, Van Hoosear, Villanueva and Walker1999).
During experimental inoculation of Tetratrichomonas preputial isolates in the cattle UGT, results have ranged from no persistence (Cobo et al., Reference Cobo, Cantón, Morrell, Cano and Campero2004) to sporadic re-detection up to 2 weeks after inoculation in mature bulls (Cobo et al., Reference Cobo, Favetto, Lane, Friend, VanHooser, Mitchell and BonDurant2007b) and no persistence beyond 6 h in young heifers (Cobo et al., Reference Cobo, Cantón, Morrell, Cano and Campero2004, Reference Cobo, Corbeil, Agnew, VanHoosear, Friend, Olesen and BonDurant2007a). Pentatrichomonas hominis derived from the cattle gut also failed to persist in the UGT of heifers (Cobo et al., Reference Cobo, Corbeil, Agnew, VanHoosear, Friend, Olesen and BonDurant2007a). Variation in results is likely due to strain and host differences, however failure of preputial Tetratrichomonas to regularly establish colonization during experimental infection (Cobo et al., Reference Cobo, Cantón, Morrell, Cano and Campero2004, Reference Cobo, Corbeil, Agnew, VanHoosear, Friend, Olesen and BonDurant2007a, Reference Cobo, Favetto, Lane, Friend, VanHooser, Mitchell and BonDurant2007b) provides strong evidence for the sporadic origin hypothesis. There was no evidence for pathology caused by any of the non-Tt. foetus trichomonads (Cobo et al., Reference Cobo, Cantón, Morrell, Cano and Campero2004).
Together, the evidence supports the hypothesis that non-Tt. foetus trichomonad isolates obtained from the cattle UGT do not represent an emerging urogenital inhabitant but rather a sporadic transmission from another source, most likely the gut. However, transmission of parasites between these mucosal sites may increase the possibility of new pathogens emerging, as has been documented for other trichomonads (Maritz et al., Reference Maritz, Land, Carlton and Hirt2014). The detection of trichomonads in the cattle UGT, and the mosaic pattern of host and mucosal site specialization amongst the trichomonads highlight their adaptability and thus zoonotic potential.
The most significant concern associated with non-Tt. foetus trichomonads in the cattle UGT is most likely misdiagnosis through confusion with Tt. foetus, which could cause unnecessary culling of suspected infected animals (Campero et al., Reference Campero, Rodriguez Dubra, Bolondi, Cacciato, Cobo, Perez, Odeon, Cipolla and BonDurant2003). The scale of misdiagnosis is unclear; the low detection frequency of trichomonads, particularly in regions which are intensely monitored, such as the UK, suggests a low frequency. However, detection rates were as high as 8.5% in some groups of bulls (Campero et al., Reference Campero, Rodriguez Dubra, Bolondi, Cacciato, Cobo, Perez, Odeon, Cipolla and BonDurant2003), suggesting the issue may be more significant in some regions. The most common method of Tt. foetus monitoring is culture-based isolation and morphological identification by light microscopy (Parker et al., Reference Parker, Campbell, McIntosh and Gajadhar2003) which may lack specificity because expertise to differentiate trichomonads based on morphology is rare (Dufernez et al., Reference Dufernez, Walker, Noël, Caby, Mantini, Delgado-Viscogliosi, Ohkuma, Kudo, Capron, Pierce, Villanueva and Viscogliosi2007). Molecular detection methods offer the advantages of improved speed and specificity (Felleisen, Reference Felleisen1997; Hayes et al., Reference Hayes, Anderson and Walker2003; Parker et al., Reference Parker, Campbell, McIntosh and Gajadhar2003) and enhanced sensitivity compared with culture-based detection (Cobo et al., Reference Cobo, Favetto, Lane, Friend, VanHooser, Mitchell and BonDurant2007b), without the need for morphological expertise. However, molecular methods are not routinely used due to high cost and impracticality in an agricultural setting. Lower cost molecular methods such as loop-assisted isothermal amplification have been applied to achieve very sensitive and specific Tt. foetus detection (Oyhenart et al., Reference Oyhenart, Martínez, Ramírez, Fort and Breccia2013) and so may offer a more practical solution. As evidence suggests Tetratrichomonas sp. are short-lived in the bull UGT (Cobo et al., Reference Cobo, Cantón, Morrell, Cano and Campero2004, Reference Cobo, Favetto, Lane, Friend, VanHooser, Mitchell and BonDurant2007b), and may originate from sexual mounting behaviour (Walker et al., Reference Walker, Hayes, Sawyer, Nordhausen, Van Hoosear and BonDurant2003), separating bulls before testing may also reduce misdiagnosis. Artificial insemination combined with regular monitoring of semen for parasites has proven a very successful control method (Dąbrowska et al., Reference Dąbrowska, Karamon, Kochanowski, Sroka, Zdybel and Cencek2019), and should also be considered for more widespread adoption.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S003118202100086X
Data
All sequences generated in this study can be obtained from the NCBI Genbank database (Benson et al., Reference Benson, Clark, Karsch-Mizrachi, Lipman, Ostell and Sayers2015) under accessions MT011380-MT011382 for the 18S rRNA sequences and MT375127-MT375129 for the ITS1-5.8S rRNA-ITS2 sequences.
Acknowledgements
We acknowledge the contribution of Alan Murphy in collecting metadata for the animal included in this study.
Author contributions
E. V. R. collected the original isolate. N. P. B. performed the molecular sequence typing, data analysis and wrote the article. R. P. H. contributed to project design and feedback, amendments and advice for writing the article.
Financial support
This study was supported by the Biotechnology and Bioscience Research Council Doctoral Training Partnership for Newcastle, Liverpool and Durham (NPB, RH, grant number: BB/M011186/1) and the Animal and Plant Health Agency (EVR).
Conflict of interest
The authors declare there are no conflicts of interest.
Ethical standards
No animals were used for experimental procedures to complete this work. The animal was sampled as part of routine, statutory sampling for bulls used in artificial insemination, as required by The Bovine Semen (England) Regulations (2007).