Introduction
Dermoergasilus Ho and Do, Reference Ho and Do1982 currently includes 12 valid species parasitizing freshwater, marine and brackish water fishes in Indian, Indo-West Pacific, Palaearctic and Afrotropic regions (Dogiel and Akhmerov, Reference Dogiel and Akhmerov1952; Cressey and Collette, Reference Cressey and Collette1970; Ho and Do, Reference Ho and Do1982; Byrnes, Reference Byrnes1986; Oldewage and Van As, Reference Oldewage and Van As1988; Ho et al., Reference Ho, Jayarajan and Radhakrishnan1992; Kabata, Reference Kabata1992; El-Rashidy and Boxshall, Reference El-Rashidy and Boxshall1999; El-Rashidy and Boxshall, Reference El-Rashidy and Boxshall2001; Hassan et al., Reference Hassan, Jones and Lymbery2009; Ali and Adday, Reference Ali and Adday2019). The host spectrum of Dermoergasilus species is broad and comprises various fishes belonging to 14 families, including mostly Mugilidae (12 species), Belonidae (4 species) and Sparidae (3 species). The number of host species parasitized by a Dermoergasilus species ranges from 1 (Dermoergasilus cichlidus Ali and Adday, Reference Ali and Adday2019, Dermoergasilus curtus El-Rashidy and Boxshall, Reference El-Rashidy and Boxshall2001 and Dermoergasilus semicoleus Cressey and Collette, Reference Cressey and Collette1970) to 8 [Dermoergasilus amplectens(Dogiel and Akhmerov, Reference Dogiel and Akhmerov1952)] (Table 1).
Table 1. Checklist of Dermoergasilus including host species, locality and site of collection

N/A, data not available.
The valid names of fish hosts are given in parentheses.
Dermoergasilus was proposed by Ho and Do (Reference Ho and Do1982) to include 3 previously described species of Ergasilus (i.e. Ergasilus amplectens Dogiel and Akhmerov, Reference Dogiel and Akhmerov1952; Ergasilus coleus Cressey and Collette, Reference Cressey and Collette1970; Ergasilus semicoleus Cressey and Collette, Reference Cressey and Collette1970) possessing a combination of the following characters: (i) antenna, except terminal claw, covered with inflated transparent membrane; (ii) paired caudal rami each with a digitiform process; and (iii) middle segment of endopod of legs II and III possessing a single seta. Later, Byrnes (Reference Byrnes1986) described Dermoergasilus acanthopagri Byrnes, Reference Byrnes1986 from sea breams (Sparidae) in Australia. Nevertheless, Gussev (Reference Gussev1987) questioned the validity of the genus when he found several Ergasilus species possessing the antennal transparent membrane. Meanwhile, Oldewage and Van As (Reference Oldewage and Van As1988) described Dermoergasilus mugilis Oldewage and Van As, Reference Oldewage and Van As1988 from grey mullet (Mugilidae) in Africa. Kabata (Reference Kabata1992) confirmed the validity of the genus and stated that even just the digitiform process on paired caudal rami distinguishes Dermoergasilus from Ergasilus. The importance of the transparent membrane on antenna is also questioned since it is not well developed at some species of Dermoergasilus, and on the contrary, there are some species of Ergasilus which have transparent inflated membrane around the antenna (e.g. E. acusicestraeus El-Rashidy and Boxshall, Reference El-Rashidy and Boxshall1999). Kabata (Reference Kabata1992) described Ergasilus intermedius Kabata, Reference Kabata1992 and stated that this species is an intermediate form between Ergasilus and Dermoergasilus, later El-Rashidy and Boxshall (Reference El-Rashidy and Boxshall1999) transferred this species to Dermoergasilus. Dermoergasilus varicoleus Ho et al. (Reference Ho, Jayarajan and Radhakrishnan1992) parasitizing Planiliza tade (Fabricius) was described in India (Ho et al., Reference Ho, Jayarajan and Radhakrishnan1992), whereas El-Rashidy and Boxshall (Reference El-Rashidy and Boxshall2001) described 3 species of Dermoergasilus from 6 species of grey mullet hosts (see Table 1): D. longiabdominalis El-Rashidy and Boxshall, Reference El-Rashidy and Boxshall2001; D. semiamplectens El-Rashidy and Boxshall, Reference El-Rashidy and Boxshall2001; and D. curtus El-Rashidy and Boxshall, Reference El-Rashidy and Boxshall2001. Dermoergasilus occidentalis Hassan et al., Reference Hassan, Jones and Lymbery2009 was described from eeltail catfishes (Plotosidae) and galaxiids (Galaxiidae) in Australia (Hassan et al., Reference Hassan, Jones and Lymbery2009). Ahmed and Ali (Reference Ahmed and Ali2013) reported Dermoergasilus sp. from common carp (Cyprinus carpio L.) in Iraq but did not provide further morphological identification. Most recently, Dermoergasilus cichlidus Ali and Adday, Reference Ali and Adday2019 was described from redbelly tilapia [Coptodon zillii (Gervais)] in Iraq (Ali and Adday, Reference Ali and Adday2019).
Until now, there are only a few parasitic crustacean records from freshwater fishes in Madagascar. Fryer (Reference Fryer1968) questioned whether it is due to the lack of scientific interest or because of their true absence. The only record of a parasitic copepod on this island is Dermoergasilus longiabdominalis El-Rashidy and Boxshall, Reference El-Rashidy and Boxshall2001 from Osteomugil engeli (Bleeker) (El-Rashidy and Boxshall, Reference El-Rashidy and Boxshall2001). From other parasitic crustaceans recorded in the region only the occurrence of parasitic isopod Cymothoa borbonica Schioedte & Meinert, 1884 from the mouth of the freshwater cichlid fish Ptychochromis oligacanthus (Bleeker) is reported by (Trilles, Reference Trilles1975).
The other parasitic crustaceans recorded from this area are associated with the marine fish species (e.g. Barnard, Reference Barnard1960; Cressey, Reference Cressey1963; Trilles, Reference Trilles1975, Reference Trilles1979, Reference Trilles2008; Benz, Reference Benz2006); or mud shrimps (Humes et al., Reference Humes, Cressey and Gooding1958); sea stars (Humes and Cressey, Reference Humes and Cressey1958; Humes and Ho, Reference Humes and Ho1966; Humes, Reference Humes1971); gorgonaceans (Humes, Reference Humes1974); holothurians (Humes and Cressey, Reference Humes and Cressey1959, Reference Humes and Cressey1961; Humes, Reference Humes1967); corals (Humes, Reference Humes1962; Humes and Frost, Reference Humes and Frost1964; Humes and Ho, Reference Humes and Ho1967); molluscs (Humes and Ho, Reference Humes and Ho1965); antipatharians (Humes, Reference Humes1967).
During the investigation of gill parasites of cichlid fishes in Madagascar, Dermoergasilus specimens were collected from the gills of Paretroplus polyactis. Description of new Dermoergasilus species was performed using morphological study (light and SEM microscopy), and a molecular study using ribosomal and mitochondrial DNA sequences (partial 18S rDNA, 28S rDNA and COI sequences). In addition, to investigate the relationship of D. madagascarensis n. sp. to other representatives of Ergasilidae, phylogenetic analyses were performed.
Materials and methods
Fish collection
During a parasitological survey in April 2016, 100 fish specimens were examined for the presence of metazoan parasites (see Supplementary Table 1) Examined fish included mainly representatives of the family Cichlidae (92 specimens), some non-cichlid fishes living in sympatry with cichlids were also examined [4 specimens of Gobiidae (Glossogobius giuris (Hamilton) and Glossogobius sp.)], 2 specimens of Mugilidae [Osteomugil robustus (Günther) and Planiliza macrolepis (Smith) and Aplocheilidae (Pachypanchax omalonotus (Duméreil))]. Fishes were sampled in 4 localities (Fig. 1): (1) Lake Ravelobe (Ankarafantsika National Park) 16°18′23.14″S–46°48′43.32″E, (2) the Anjingo River (near Antsohihy) 14°50′40.89″S–48°14′43.36″E, (3) the crater lakes of Mont Passot (on Nosy Be Island) 13°19′1.84″S–48°14′3.60″E, and (4) the Canal des Pangalanes (at Andevoranto) 18°57′17.50″S–49°6′29.90″E. These areas belong to the eastern basins and freshwater systems of north-western Madagascar, all recognized as hotspots of Malagasy fish diversity (Benstead et al., Reference Benstead, De Rham, Gattolliat, Gibon, Loiselle, Sartori, Sparks and Stiassny2003). All fish specimens were transported alive to the field laboratory, sacrificed by severing the spinal cord, and dissected within 48 h following classical parasitological dissection procedure (Ergens and Lom, Reference Ergens and Lom1970). Fish specimens were measured and identified by local co-workers familiar with the fish fauna, and the identification was subsequently confirmed using sequences of the cytochrome mitochondrial gene (see Šimková et al., Reference Šimková, Řehulková, Rasoloariniaina, Jorissen, Scholz, Faltýnková, Mašová and Vanhove2019 for detailed information). The present study was part of a larger investigation concerning transmission of parasites from introduced cichlids to native Malagasy fish (Šimková et al., Reference Šimková, Řehulková, Rasoloariniaina, Jorissen, Scholz, Faltýnková, Mašová and Vanhove2019).

Figure 1. Map of Madagascar indicating the sampling localities: (1) Lake Ravelobe; (2) Anjingo River; (3) crater lakes of Mont Passot; (4) Canal des Pangalanes.
Parasite collection and identification
Live copepods were collected from the gills using fine needles and processed for morphological and molecular purposes, as described in Míč et al. (Reference Míč, Řehulková and Seifertová2023). The mounted specimens in GAP (mixture of glycerine and ammonium picrate) or pure glycerine were studied using an Olympus BX61 microscope equipped with phase contrast optics. Drawings of the copepods were made using an Olympus drawing attachment and edited with a graphic tablet (Wacom Intuos5 Touch) compatible with Adobe Illustrator and Adobe Photoshop (Adobe Systems Inc., San Jose, CA, USA). All measurements (in micrometers) were taken using digital image analysis software (Olympus Stream Motion v. 1.9.3) and are presented as the range followed by the mean (n = 10).
For scanning electron microscope analysis, 5 specimens fixed in 70% ethanol were dehydrated in an increasing ethanol grades, dried in a CPD 030 critical point drying apparatus (Bal-tec, Balzers, Liechtenstein) using liquid CO2, mounted on aluminium stubs with double sided adhesive discs, coated with gold in a SCD 040 sputter coating unit (OC Oerlikon Balzers Coating, Balzers, Liechtenstein) and examined in a VEGA scanning electron microscope operating at 20 kV.
For comparative purposes, specimens of the following 4 previously described species of Dermoergasilus available in the Natural History Museum (London, UK; BMNH) were examined: D. amplectens (BMNH 1999.1399-1401), D. longiabdominalis (BMNH 1999.1321), D. semiamplectens (BMNH 1999.1341-1374; BMNH 1999.1376-1377) and D. varicoleus (BMNH 1999.1412-1417).
The type specimens of the copepods collected in the present study were deposited in the Institute of Parasitology, Czech Academy of Sciences, České Budějovice, Czech Republic. Prevalence (percentage of infected fish) and mean intensity of infection (mean number of parasites per infected host) were calculated following Bush et al. (Reference Bush, Lafferty, Lotz and Shostak1997).
Molecular and phylogenetic analyses
Genomic DNA was isolated separately from each parasite specimen (or a part of its body) using DNeasy®Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. For molecular characterization, partial sequences of 1 mitochondrial gene (COI) and 2 nuclear ribosomal regions (18S and 28S rDNA) were amplified by using the primer sets listed in Table 2. PCRs for 18S and 28S rDNA were carried out in a total volume of 20 μL containing 3 μL of DNA extract, 1× PCR buffer (Fermentas), 2.5 mm MgCl2, 0.2 mm of each dNTP, 0.2 μ m of each primer, 0.1 BSA and 1 U of Taq polymerase (Fermentas). Amplification was performed under the following conditions: 94°C for 5 min; 39 cycles of 94°C for 30 s; an annealing temperature of 52°C for 30 s; and 72°C for 1 min, with a final extension step at 72°C for 5 min. PCR for COI was carried out in a total volume of 50 μL containing 1 μL of DNA extract, 1× PCR buffer (Fermentas), 2.5 mm MgCl2, 0.5 mm of each dNTP, 0.5 μ m of each primer, 0.1 BSA and 2 U of Taq polymerase (Fermentas). Amplification was performed under the following conditions: 95°C for 5 min; 40 cycles of 95°C for 1 min; an annealing temperature of 45°C for 1 min; and 72°C for 30 s, with a final extension step at 72°C for 7 min. The PCR amplicons were checked by electrophoresis on 1.5% agarose gels stained with Good View™ (Amplia s.r.o., Bratislava, Slovakia), and PCR products of the required length were purified using ExoSAP-IT™ (Affymetrix Inc., Santa Clara, USA), following the manufacturer's instructions. Purified products were directly sequenced using the same primers as those for PCR. DNA sequencing was carried out using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems by Thermo Fisher Scientific, Prague, Czech Republic) and a 3130 Genetic Analyzer (Applied Biosystems). The obtained sequences were assembled and edited using Sequencher software (Gene Codes Corp., Ann Arbor, MI, USA). Newly generated sequences of 18S rDNA, 28S rDNA and COI were deposited in GenBank under accession numbers PP115569 (28S rDNA), PP115568 (18S rDNA) and PP117929-PP117934 (COI). Molecular vouchers (hologenophores, paragenophores; Pleijel et al., Reference Pleijel, Jondelius, Norlinder, Nygren, Oxelman, Schander, Sundberg and Thollesson2008) were deposited in the Institute of Parasitology, Czech Academy of Sciences, České Budějovice, Czech Republic.
Table 2. List of primers used for PCR amplifications of mitochondrial and nuclear markers in the present study

Ta, annealing temperature.
To investigate the phylogenetic position of Dermoergasilus madagascarensis n. sp., to the representatives of parasitic Cyclopoida, the sequences of 28S rDNA of the species belonging to 9 genera were retrieved from GenBank and Bold databases (for details, see Table 3). Three species of the family Lernaeidae, Lernaea cyprinacea (Linnaeus, 1758), Lamproglena chinensis Yü, 1937 and Lamproglena orientalis Markevich, 1936 were used as outgroup. Sequences were aligned using MAFFT v.7 (Katoh and Standley, Reference Katoh and Standley2013). Gaps and ambiguously aligned regions were removed from the alignments with Gblocks v0.91b (Talavera and Castresana, Reference Talavera and Castresana2007) using settings for a less stringent selection. ModelFinder (Kalyaanamoorthy et al., Reference Kalyaanamoorthy, Minh, Wong, Von Haeseler and Jermiin2017) was employed to select the most appropriate model of DNA evolution. The most suitable evolutionary model for the partial sequence of 28S rDNA was TIM3 + F + I. The phylogenetic reconstruction was performed using maximum likelihood (ML) and Bayesian inference (BI) methods. ML analyses were run using IQ-TREE (Nguyen et al., Reference Nguyen, Schmidt, von Haeseler and Minh2015) on the W-IQ-TREE webserver (Trifinopoulos et al., Reference Trifinopoulos, Nguyen, von Haeseler and Minh2016) and nodal support for the tree was assessed through ultrafast bootstrap approximation with 1000 replicates (Hoang et al., Reference Hoang, Chernomor, von Haeseler, Minh and Vinh2018). BI analysis was carried out in MrBayes 3.2.6 (Huelsenbeck and Ronquist, Reference Huelsenbeck and Ronquist2001) using the CIPRES platform (Miller et al., Reference Miller, Pfeiffer and Schwartz2010), the analysis included 2 simultaneous runs of Markov chain Monte Carlo for 106 generations, sampling every 100 generations, with a ‘burn-in’ of 25%. The results were checked in Tracer v. 1.7.1 (Rambaut et al., Reference Rambaut, Drummond, Xie, Baele and Suchard2018) to assess chain convergence. The trees were visualized and edited in FigTree v. 1.4.3 (Rambaut, Reference Rambaut2012). Genetic distances (uncorrected p-distance) were calculated in MEGA v. 11 (Tamura et al., Reference Tamura, Stecher and Kumar2021).
Table 3. List of parasitic copepods used for phylogenetic analyses and calculation of p-distances, including their host species, collection locality, and accession numbers for partial 18S, 28S rDNA and COI sequences from database GenBank and Bold (indicated with *)

Newly generated sequence is given in bold.
Results
Endemic cichlid P. polyactis from the Canal des Pangalanes (locality 4 in Fig. 1) was the only host species (out of 15 species examined) infected by parasitic copepods and exhibited intensity of infection ranging from 5 to 283 (mean 59) per individual fish. Overall, 20 specimens of P. polyactis were examined and the prevalence of Dermoergasilus parasites was 90%. Total prevalence of Dermoergasilus among all examined fishes in the study was 18%.
The copepod specimens collected from P. polyactis were identified as Dermoergasilus based on the diagnostic morphological characters according to Ho and Do (Reference Ho and Do1982), specifically: (i) antenna, except terminal claw, covered with inflated transparent membrane; (ii) paired caudal rami each with a digitiform process; and (iii) middle segment of endopod of legs II and III possessing a single seta.
Family Ergasilidae Burmeister, 1835
Genus Dermoergasilus Ho & Do, 1982
Dermoergasilus madagascarensis n. sp.
Type-host: Paretroplus polyactis (Bleeker, 1878) (Cichlidae, Cichliformes)
Type-locality: Canal des Pangalanes (at Andevoranto) (18°57′17.50″S, 49° 6′29.90″E), Madagascar
Type and voucher material: Holotype (adult female): Cr-39 (1 specimen). Paratypes (adult females): Cr-39 (3 specimens). Hologenophores (adult females): Cr-39 (16 specimens).
Site on host: Gill filaments.
Prevalence and intensity of infection: 90% (18 fish infected/20 fish examined); 5–283 (mean 59) copepods per infected host.
ZooBank registration: urn:lsid:zoobank.org:act:5A5C2DCB-CCAB-4545-B6F4-F416CC22B10D
Representative DNA sequences: A 1384 bp long 18S rDNA sequence, 674 bp long 28S rDNA sequence and 9 COI sequences of 678 bp long obtained from 10 specimens are deposited in the NCBI GenBank database under the accession numbers PP115569 (28S rDNA), PP115568 (18S rDNA) and PP117929-PP117934 (COI), respectively.
Etymology: The species was named after the type locality, Madagascar Island, from which it was first discovered.
Description
Adult female. [Based on 10 specimens; Figs 2–5; measurements in Table 4].

Figure 2. Dermoergasilus madagascarensis n. sp., adult female from Paretroplus polyactis. (A) habitus, dorsal; (B) antenna, ventral; (C) mandible and maxilulle, ventral (D) maxilla, ventral; (E) antennule, ventral.

Figure 3. Dermoergasilus madagascarensis n. sp., adult female from Paretroplus polyactis. (A) abdomen and caudal rami; (B) egg sac, dorsal; (C) leg 5, ventral; (D) rostrum, dorsal; (E) interpodal plates, ventral.

Figure 4. Dermoergasilus madagascarensis n. sp., adult female from Paretroplus polyactis. (A) leg 1, ventral; (B) leg 2, ventral; (C) leg 3, ventral; (D) leg 4, ventral.

Figure 5. Scanning electron micrographs of Dermoergasilus madagascarensis n. sp., adult female from Paretroplus polyactis. (A) entire female body, carrying egg sacs, lateral; (B) antenna with transparent membrane (arrow), dorsal; (C) cephalosome with sensory setae and pits (arrow); antennule, lateral dorsal; (D) interpodal plates with ornamentation (arrow), ventral; (E) caudal rami and digitiform process (arrow), ventral; (F) leg 5, ventral.
Table 4. Measurements (in micrometers) of specimens (n = 10) of Dermoergasilus madagascarensis n. sp. parasitizing endemic cichlid Paretroplus polyactis in Madagascar

Prosome 5-segmented, composed of cephalothorax and 3 free pedigerous somites (PS-1 to PS-4) (Fig. 2A). Cephalosome roughly pentagonal, rounded and slightly tapering anteriorly; antennules and antennae visible in dorsal view (Fig. 5A and B). Cephalic ornamentation comprising inverted T-shaped marking, sensory setae and pits with bilaterally symmetrical distribution on dorsal side. Rostrum shieldlike with 6 sensillae and 3 integumental pores (Figs. 3D and 5C). PS-1 elongated, with bilateral indentations just posterior to midlength; dorsal surface with slight T-shaped and rectangular depression situated anterior and posterior, respectively, to the constricted part; dorsal ornamentation comprising circular indentations situated just posterior to cephalosome and pair of sensillae near posterior margin. PS-2 to PS-4 decreasing gradually in width posteriorly, the three together barrel-shaped. Dorsal surface of each segment possessing anteriorly arising trapezoidal plate, sensillae and pits with bilaterally symmetrical distribution.
Urosome comprising fifth pedigerous somite (PS-5), genital double somite, and 3 free abdominal somites (AS-1 to AS-3) (Fig. 3A). PS-5 reduced, smaller and thinner than prosome somites, unornamented. Genital segment large, barrel-shaped, with transverse row of spinules and pair of hook-shaped ornamentation on ventral side. Free abdominal somites decreasing in width posteriorly. AS-1 wider than long (1.2–1.3 times), almost 3 times larger than AS-2, bearing transverse row of spinules at widest part. AS-2 slightly larger than AS-3, with transverse row of spinules at midlength. AS-3 (anal somite) deeply incised posteromedially, with spinules on posterior margin.
Caudal rami nearly equal in length with AS-3, slightly wider than long; each projecting into tapering digitiform process (about 1.6 times longer than body of ramus) with inconspicuous terminal seta (Fig. 5E) and bearing 3 terminal setae – the innermost longest and thickest, ornamented with transversal rings of inconspicuous scales at posterior 3/4; 2 lateral setae longer than digitiform processes. Two cylindrical egg-sacs, much longer than wide (4 times), each composed of 2–4 rows of eggs (Fig. 3B).
Antennule (Figs 2E and 5C) 6-segmented, tapering, distally armed with simple setae; setal formula from proximal to distal segments: 3–9 – 5–4 + ae – 2 + ae – 7 + ae. Antenna (Figs 2B and 5B) comprising coxobasis, 3-segmented endopod (Enp-1 to Enp-3), and strongly recurved terminal claw. Enp-1 (proximal) longest, nearly 1.7 times longer than coxobasis, slightly inflated medially, unornamented; Enp-2 (medial) elongated, slightly curved, about half length of Enp-1, unornamented; ES-3 inconspicuous, unornamented. Terminal claw curved, about half size of ES-2, with inconspicuous subterminal inner denticle. Antenna (except terminal claw) covered with inflated cuticular membrane, without setules, spines or indentations.
Mouthparts (Fig 2C and D) comprising mandible, maxillule and maxilla; maxilliped absent. Mandible consisting of 3 blades (anterior, middle and posterior); anterior blade with sharp teeth on anterior margin; middle blade with sharp teeth on both margins; and posterior blade with sharp teeth on anterior margin. Maxillule a single lobe, ornamented with rows of tiny spinules, bearing 2 equally long outer setae and minute inner seta. Maxilla 2-segmented, comprising syncoxa and basis; syncoxa small, unarmed; basis elongated, medially slightly curved, distally with numerous sharp teeth on anterior side.
Swimming legs (L1–L4) biramous; each comprising coxa, basis, endopod (inner ramus), and exopod (outer ramus) (Fig. 4). Intercoxal sclerites slender; each with tapering ends directed posterolaterally, unornamented. Interpodal plates slender, uniform in shape; each with 2 inconspicuous bilateral pores and 3–4 transversal rows of spinules (Figs 3E and 5D). Armature formula of L1–L4 (spines – Roman numerals; setae – Arabic numerals) shown in Table 5.
Table 5. Spine (Roman numerals) and setal (Arabic numerals) formula of swimming legs of D. madagascarensis n. sp.

Coxa of all legs unarmed; coxa of L1 with a row of spinules extending along its outer posterior margin. Basis of all legs armed with proximal outer spine, unornamented. Legs 1–4 with outer margin of both rami ornamented with rows of spinules; outer and inner margin of first endopodal and exopodal segment, respectively, of all legs partly or completely covered with bristles.
Leg 1 (Fig. 4A): exopod 3-segmented; first segment with small naked spine arising from outer posterior margin; second segment with inner plumose seta; third segment with 2 blade-like serrated spines (shorter more proximal), 1 semi-plumose seta (=seta with outer margin serrated) and 4 plumose setae.
Endopod 3-segmented; first and second segment each with 1 plumose seta; third segment with 3 plumose setae, 1 semi-plumose seta and 2 blade-like serrated spines.
Leg 2 (Fig. 4B): exopod 3-segmented; first segment with small outer spine; second segment with 1 plumose seta; third segment with 1 semi-plumose seta and 5 plumose setae.
Endopod 3-segmented; first and second segments each with 1 small slender serrated spine, 1 plumose seta; third segment with 3 plumose setae, 1 semi-plumose seta.
Leg 3 (Fig. 4C): exopod 3-segmented; first segment with small outer spine; second segment with 1 plumose seta; third segment with 1 semi-plumose seta and 5 plumose setae. Endopod 3-segmented; first and second segments each with 1 plumose seta; third segment with 1 small slender serrated spine, 3 plumose setae and 1 semi-plumose seta.
Leg 4 (Fig. 4D): exopod 2-segmented; first segment elongated, with small outer spine; second segment with 5 plumose setae. Endopod 3-segmented; first segment with 1 plumose seta; second segment with 2 plumose setae; third segment with 1 slender serrated spine and 3 plumose setae.
Leg 5 (Figs 3C and 5F): reduced but clearly visible, 2-segmented. Basal segment very small and visible dorsally, bearing outer seta; distal segment with 3 setae on inner margin (apical seta largest).
Specimens preserved in ethanol faint brown in colour, with blue spot in eyespot and sometimes in cephalothorax.
Male: Unknown
Remarks
Dermoergasilus madagascarensis n. sp. represents another species of Dermoergasilus, besides D. curtus (El-Rashidy and Boxshall, Reference El-Rashidy and Boxshall2001) and D. intermedius (Kabata, Reference Kabata1992), that have antennae with only slightly inflated cuticular membrane. All other known Dermoergasilus spp. possess a conspicuous balloon-like inflated membrane covering all or only the first (in D. semicoleus) antennal endopodal segment. In D. curtus, however, the cuticular membrane covers only the inner surface of the first endopodal segment of the antenna, whereas in D. intermedius and D. madagascarensis n. sp. the membrane ensheathes all endopodal segments. The new species differs further from D. curtus mainly by having: (i) a pentagon-shaped cephalosome (vs bullet-shaped cephalosome); (ii) second endopodal segment of the antenna without a minute seta (vs with a minute seta proximally on inner margin of the segment); (iii) interpodal plates ornamented with 3–4 rows of spinules (vs 1 row of spinules); (iv) genital segment with 1 medial row of spinules (vs 3 posterior rows of spinules); (v) urosomites without folded membrane; and (vi) 2 lateral caudal setae longer than the digitiform process (vs shorter than the digitiform process). Dermoergasilus madagascarensis n. sp. is easily differentiated from D. intermedius by having: (i) anteriorly rounded and slightly tapering pentagonal cephalosome (vs anteriorly flat square-shaped cephalosome with widely separated antennules); (ii) second endopodal segment of the antenna medially swollen (vs the segment slender and of the same diameter along entire antenna); (iii) interpodal plates ornamented with 3–4 rows of spinules (vs unornamented); (iv) genital segment with 1 medial row of spinules (vs 1 posterior row of spinules, sometimes with gaps in middle part); (v) 2 lateral caudal setae longer than the digitiform processes (vs 1 longer and 1 shorter than the digitiform process); and (vi) a different armature formula of the third endpodal segment of legs II to IV.
In terms of the armature of the swimming legs, D. madagascarensis n. sp. shares the same spine and setal formula with 6 other species of Dermoergasilus, namely D. amplectens, D. cichlidus, D. curtus, D. longiabdominalis, D. occidentalis and D. semiamplectens, recorded on fishes of different families, but mostly of the Mugilidae (see Table 1). With the exception of D. curtus, all 5 species mentioned above are clearly distinguished from the new species by having a slender urosomite (genital segment and the first abdominal somite are markedly elongated vs both barrel shaped in D. madagascarensis n. sp.).
Dermoergasilus madagascarensis n. sp. is the first recorded copepod parasitizing freshwater fishes in Madagascar and besides D. amplectens from orange chromid Pseudetroplus maculatus (Bloch) (India; Ho et al., Reference Ho, Jayarajan and Radhakrishnan1992) and D. cichlidus from Coptodon zilii (Iraq; Ali and Adday, Reference Ali and Adday2019), it is the third species of Dermoergasilus hitherto recorded from the gills of a cichlid fish.
Molecular characterization and phylogenetic position of D. madagascarensis n. sp. within the Ergasilidae
Partial fragments of 18S (1384 bp), 28S (674 bp) rDNA and COI (675 bp) were obtained from 10 individuals of D. madagascarensis n. sp. No intraspecific sequence variability was found for any of nuclear ribosomal markers (partial 18S and 28S rDNA). Six haplotypes were found in the COI mtDNA with a low intraspecific genetic variation of 0.15–1.48%. Genetic comparison of D. madagascarensis n. sp. with other Ergasilidae species showed the lowest interspecific genetic distance with Ergasilus megaceros Wilson, 1916 (17.7%) and highest interspecific genetic distance with Neoergasilus japonicus (Harada, 1930) (23.9%) for COI sequences (Table 6). When comparing D. madagascarensis n. sp. to other ergasilid species in rDNA sequence data, the minimum interspecific distances were observed with E. sieboldi von Nordmann, 1832 (0.9% for 18S rDNA, and 4.3% for 28S rDNA) and maximum interspecific divergences were observed with Therodamas longicollum Oliveira et al. (Reference Oliveira, Corrêa, Adriano and Tavares-Dias2021) (3.4% for 18S rDNA) and Sinergasilus major (Markevich, 1940) (10.9% for 28S rDNA).
Table 6. Interspecific genetic variabilities of family Ergasilidae

Below the diagonal are showed the values for 18S rDNA (first line) and 28S rDNA(second line) and above the diagonal for COI. The range indicates minimum and maximum value of the genetic variability for species of the genus. Numbers in brackets indicate the number of species with available sequences for the specific marker (18S, 28S, COI). Bold numbers only indicate values for the genus Dermoergasilus, which is the focus of this article.
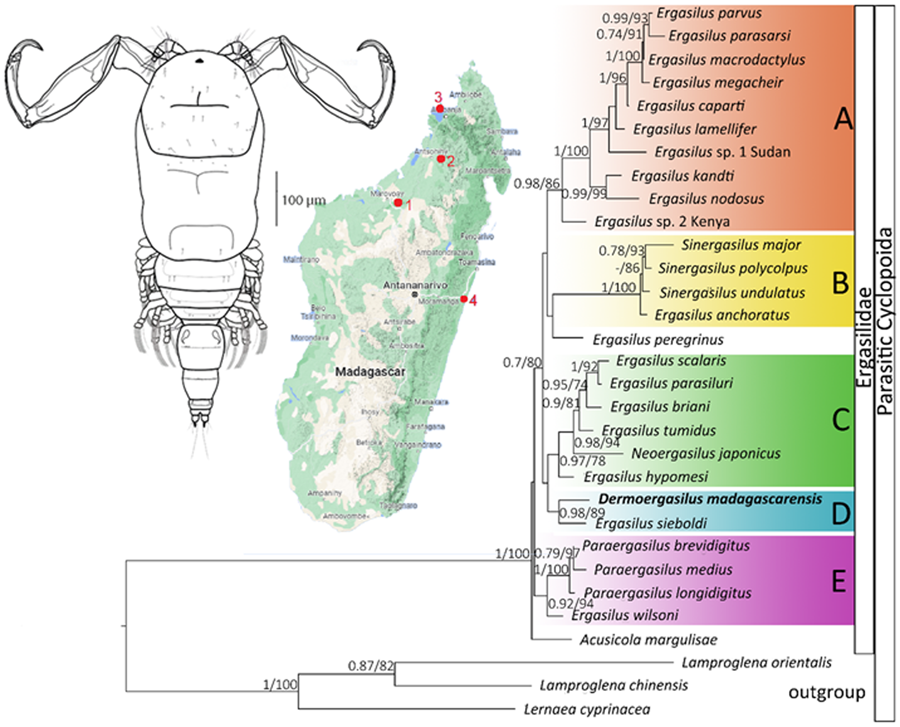
ML and BI analyses based on 28S rDNA sequences of Ergasilidae yielded trees with congruent topologies with similar nodal support values and revealed 5 well-supported groups (Fig. 6): (A) African Ergasilus species group; (B) Asian Sinergasilus species and the Ergasilus anchoratus Markevich, 1946 group; (C) Asian Ergasilus species and the Neoergasilus japonicus group, (D) E. sieboldi and D. madagascarensis n. sp. group and (E) Paraergasilus species and the Ergasilus wilsoni Markevich, 1933 group. Phylogenetic reconstruction showed the polyphyletic status of the genus Ergasilus.

Figure 6. Phylogenetic tree of Ergasilidae reconstructed by Maximum Likelihood. The tree is based on the partial 28S rDNA sequences (674 bp alignment). Values along the branches indicate posterior probabilities from Bayesian Inference and bootstrap values from Maximum Likelihood (dashes indicate values below 0.7 and 50, respectively).
Discussion
Diversity of fish ectoparasites in native Malagasy freshwater fish has been little studied in the past. The present study was a part of large parasitological investigation performed only in 4 localities of north-western Madagascar, however, documenting unknown diversity of fish parasites in isolated freshwater region with endemic fish fauna (i.e., Madagascar), the pattern which was previously shown for endemic freshwater fish in other regions i.e., Peri-Mediterranean and Middle East (Benovics et al., Reference Benovics, Kičinjaová and Šimková2017, Reference Benovics, Koubková, Civáňová, Rahmouni, Čermáková and Šimková2021; Rahmouni et al., Reference Rahmouni, Řehulková, Pariselle, Rkhami and Šimková2017; Řehulková et al., Reference Řehulková, Benovics and Šimková2020; Nejat et al., Reference Nejat, Benovics, Řehulková, Vukić, Šanda, Kaya, Tarkan, Abdoli, Aksu and Šimková2023). Prior to this study, 12 valid species of Dermoergasilus were known, including 1 species, specifically D. longiabdominalis, in mugilid hosts in Madagascar. Two Dermoergasilus species were previously reported on cichlid hosts in India and Iraq. The first species, D. amplectens, was recorded on a number of fish species and over a wide geographic range, including Pseudetroplus maculatus, an endemic cichlid of southern India and Sri Lanka. The second species, D. cichlidus, was described from Coptodon zillii, a non-native cichlid in Iraq. Dermoergasilus madagascarensis n. sp. represents the third species of the genus reported on cichlids and the second species of the genus revealed in Madagascar and a single known species currently known only from endemic Malagasy cichlids (i.e., P. polyactis).
Even though questioned in the past (Gussev, Reference Gussev1987; Kabata, Reference Kabata1992; El-Rashidy and Boxshall, Reference El-Rashidy and Boxshall2001), Dermoergasilus still remains valid. From the 3 morphological characters proposed by Ho and Do, Reference Ho and Do1982 only 1 clearly differentiates this genus, which is a digitiform process on each paired caudal rami. The other 2 characters seem to be ambiguous. The inflated transparent membrane is quite a vague morphological character, and some species of Dermoergasilus do not have it well developed (e.g. D. curtus or D. intermedius). The membrane could be an ancestral trait that is being lost during the evolution, from clearly visible balloon-like inflation in D. amplectens to barely noticeable cuticle in D. curtus. Moreover, there are some Ergasilus species with some kind of hyaline membrane on antenna. For example, the membrane on antenna of Ergasilus megacheir (Sars, 1909) appears to be very similar to that of D. curtus. The middle segment of endopod of legs II and III possessing a single seta is even less persuasive character, since at least 10 Ergasilus species (e.g. E. tumidus Markevich, 1940, E. briani Markevich, 1933, E. gibbus von Nordmann, 1832, E. gobiorum Markevich & Sukhnenko, 1967 etc.) also possess this character (Ho et al., Reference Ho, Jayarajan and Radhakrishnan1992; Kabata, Reference Kabata1992). There are other morphological traits present in most of the species of Dermoergasilus, e.g. long first free abdominal segment, similar morphology of leg 5, falciform seta on legs, some species even share the same spine-seta and antennal formula. However, neither of them can clearly distinguish Dermoergasilus from other members of Ergasilidae but could indicate their possible close relationship and a common ancestry.
Based on the literature review, D. madagascarensis n. sp. shares the same spine and setal formula with 6 other species of the genus. Future studies using molecular analyses should focus on this aspect and verify, if species with the same armature of swimming legs are phylogenetically related. Many of these species were recorded from mugilid hosts in Indian region. It is possible that they have the common origin, and the divergence of the species is associated with geographical isolation of Madagascar, drifting away from the Indian peninsula 96–65 Mya (Vences et al., Reference Vences, Wollenberg, Vieites and Lees2009). El-Rashidy and Boxshall (Reference El-Rashidy and Boxshall2001) suggested that a mugilid as a host is a plesiomorphic character for Dermoegasilus, and that the ancestor of this group of parasites also occurred on a mugilid host. Acquiring hosts of other fish families could be a result of the adaption to the conditions in the new environment, which are cichlids in this case. However, only molecular data from D. curtus, D. longiabdominalis and D. semiamplectens reported from mugil hosts in India, China, Madagascar, Philippines and Myanmar would shed more light on the origin of D. madagascarensis n. sp. and clarify its relationship with other Dermoergasilus and Ergasilus species.
Scanning electron microscopy (SEM) is the method providing the appropriated visualization of some morphological structures, in our study, specifically sensory setae and pits, and also the minute seta on the digitiform process in D. madagascarensis n. sp., while the latter character was not visible under the light microscope. It is highly likely that some morphological characters might be overlooked in older descriptions of Ergasilidae, in which authors did not use SEM.
The present results of phylogenetic analyses are consistent with previously reported ergasilid phylogenies (Song et al., Reference Song, Wang, Yao, Gao and Nie2008; Santacruz et al., Reference Santacruz, Morales-Serna, Leal-Cardín, Barluenga and Pérez-Ponce de León2020; Kvach et al., Reference Kvach, Tkachenko, Seifertová and Ondračková2021; Míč et al., Reference Míč, Řehulková and Seifertová2023). Phylogenetic reconstruction based on 28S rDNA presented in this study showed the sister relationship among newly described D. madagascarensis n. sp. and E. sieboldi von Nordmann, 1832, a cosmopolitan parasite of freshwater fishes (Yamaguti, Reference Yamaguti1939; Kabata, Reference Kabata1979; Amado et al., Reference Amado, Falavigna da Rocha, Piasecki, Al-Daraji and Mhaisen2001). While the cephalothorax shape is similar between the 2 species, the new species differs from E. sieboldi by: (i) digitiform process on caudal rami (ii) absence of spines on antenna (vs short spine on inner surface of the first endopodal segment of antenna and 2 short spines on inner surface of the second endopodal segment of antenna in E. sieboldi); (iii) absence of circular structure posterior to inverted T-structure on cephalothorax (vs presence in E. sieboldi), (iv) caudal rami bearing 3 terminal setae (vs 4 terminal setae in E. sieboldi); (v) having only 1 seta on the second segment of the endopods of legs II and III (vs 2 setae in E. sieboldi).
However, we can still ask whether the position of D. madagascarensis n. sp. in the phylogenetic tree is because of the real relatedness of these 2 species or due to the lack of molecular data for other species of the family Ergasilidae, especially those currently included in Dermoergasilus. A close relationship among D. madagascarensis n. sp. and African species of Ergasilus has not been confirmed in present study, so the newly described species does not appear to originate from Africa (at least based on the phylogeny including currently available DNA sequences of African Ergasilus). A fragment of COI mtDNA gene was also successfully obtained for representative number of D. madagascarensis n. sp. specimens. Unfortunately, no other DNA data are currently available for representatives of the Dermoergasilus genus and no threshold for intra- or interspecific variability was set for ergasilid species. However, the distances between COI haplotypes of D. madagascarensis n. sp. did not exceed 1.5%, the intraspecific limit generally accepted for COI mtDNA of Copepoda (Bucklin et al., Reference Bucklin, Frost, Bradford-Grieve, Allen and Copley2003; Dippenaar et al., Reference Dippenaar, Mathibela and Bloomer2010; Laakmann et al., Reference Laakmann, Gerdts, Erler, Knebelsberger, Martínez Arbizu and Raupach2013). The COI intraspecific distances in other ergasilid species reached the values from 0% (E. wilsoni or E. jaraquensis Thatcher & Robertson B.A., 1982) to 6.9% (N. japonicus). In contrast, COI distances between Dermoergasilus and other genera reached values over 17%, supporting it being a separate genus. Nevertheless, to clearly resolve the phylogeny of Ergasilidae, DNA sequences of more ergasilid species from other parts of the world are needed.
Conclusion
Based on morphological and molecular data, a new species of Dermoergasilus has been described. Dermoergasilus madagascarensis n. sp. from the cichlid P. polyactis is the second report of a representative of the genus in Madagascar and the first molecular data for the genus were obtained. Even though the validity of the genus was questioned in the past, the possession of digitiform process on caudal rami clearly distinguishes it from other genera of the Ergasilidae. However, our phylogenetic analyses showed the polyphyly of the genus Ergasilus, and the close phylogenetic relationship between D. madagascarensis n. sp. and widely geographically distributed Ergasilus sieboldi. We highlight that more molecular data are needed to clarify the relationships between the species of Dermoergasilus and their position within the Ergasilidae.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182024000088.
Data availability statement
Type and voucher specimens were deposited in the Institute of Parasitology, Czech Academy of Sciences, České Budějovice, Czech Republic (accession code Cr-39). The sequences produced in this study were deposited in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ (accession codes PP115568, PP115569, PP117929-PP117934).
Acknowledgements
The authors are grateful to M. P. M. Vanhove (former employee of Masaryk University, Czech Republic, currently Hasselt University, Belgium) for his participation on fish dissection and parasite collection during the field trip realized in Madagascar in 2016. Our special thanks go to Jean Robertin Rasoloariniaina, Roger Daniel Randrianiana, Sarah Rakotomamonjy and Natacha Rasozolaka (University of Antananarivo) and the Centre National de Recherches Océanographiques at Nosy Be for help in the field; Lance Woolaver (Durrell Wildlife Conservation Trust) for enabling the use of the laboratory in Ankarafantsika National Park; Leonel Angelier Jaofeno for authorizing fishing in the Mont Passot Site lakes; Sylvère Lalao Rakotofiringa for advice; Joël Ho Shing Lone for the possibility to use the field laboratory. We thank Iveta Hodová and Naděžda Vaškovicová (Masaryk University, Czech Republic) for their help with SEM. We also thank Miranda Lowe and Donney Nicholson (Natural History Museum, London, United Kingdom) for the kind loan of type specimens and comparative material.
Authors’ contributions
AŠ, RM and MS conceived and designed the study. EŘ and AŠ collected parasitological material, RM performed morphological characterization and described the species. RM and MS performed molecular and phylogenetic analyses. RM and MS wrote the first draft of the manuscript, EŘ reviewed description of the species. All authors substantially contributed to the final draft and approved the final version of the manuscript.
Financial support
This study was financially supported by the Czech Science Foundation (Project No. P505/12/G112). RM was supported by the Masaryk University, Czech Republic, project no. MUNI/A/1422/2022.
Competing interests
None.
Ethical standards
All applicable institutional, national and international guidelines for the care and use of animals were followed. The fish sampling was carried out following permission No. 06/AR.ED./15 issued on April 1, 2016 by the General Directorate for Fishery Resources and Fisheries, Ministry of Fisheries Resources and Fisheries, Madagascar.