Introduction
Cosmocercoides Wilkie, 1930 are gastrointestinal parasites commonly found in amphibians and reptiles, and occasionally in terrestrial snails and slugs. These nematodes are characterized mainly by the rosette papillae on the male caudal region (Chen et al., Reference Chen, Zhang, Nakao and Li2018a; Liu et al., Reference Liu, Yu, Shu, Zhao, Fang and Wu2019; Dos Anjos et al., Reference Dos Anjos, Oda, Campião, Ávila, Dos Santos, Dos Santos, Almeida, Melo and Rodrigues2021). The species have monoxenic life cycle and adult females release eggs in host feces that develop into first-stage larvae in the environment. The larvae moult twice and become an infective third-stage larva that penetrates through the skin of a new host (Anderson, Reference Anderson2000).
Currently, there are approximately 28 nominal species of Cosmocercoides distributed worldwide. Of those, only 4 species have been reported from the Neotropical region, including Cosmocercoides lilloi Ramallo et al., Reference Ramallo, Bursey and Goldberg2007 from Rhinella arenarum (Hensel, 1867); Cosmocercoides latrans Draghi et al., Reference Draghi, Drago and Lunaschi2020 from Leptodactylus luctator (Hudson, 1892), both from Argentina; Cosmocercoides sauria Ávila et al., 2010 from lizard Iphisa elegans (Gray, 1851) and Cosmocercoides meridionalis Anjos et al., 2021 from Boana geographica (Spix, 1824), Boana boans (Linnaeus, 1758), Dryaderces cf. inframaculata (Boulenger, 1882), Osteocephalus taurinus (Steindachner, 1862) and Phyllomedusa camba (De la Riva, 1999), both from Brazil (Ramallo et al., Reference Ramallo, Bursey and Goldberg2007; Ávila et al., Reference Ávila, Strüssmann and Da Silva2010; Draghi et al., Reference Draghi, Drago and Lunaschi2020; Dos Anjos et al., Reference Dos Anjos, Oda, Campião, Ávila, Dos Santos, Dos Santos, Almeida, Melo and Rodrigues2021).
Until now, molecular data available for Cosmocercoides spp. are very scarce, and only Cosmocercoides tonkinensis Tran, Sato and Luc, Reference Tran, Sato and Luc2015; Cosmocercoides qingtianensis Chen et al., Reference Chen, Zhang, Nakao and Li2018; Cosmocercoides wuyiensis Liu et al., 2019; Cosmocercoides pulcher Wilkie, Reference Wilkie1930 from Oriental region and Cosmocercoides dukae Holl, 1928 from Nearctic region were studied using molecular tools (Chen et al., Reference Chen, Zhang, Nakao and Li2018a, Reference Chen, Zhang, Feng and Li2020). Thus, we used an integrative approach, including light and scanning electron microscopy and molecular analysis, to describe and characterize a new species of Cosmocercoides and determine the phylogenetic position of this species.
Materials and methods
Host collection and morphological study of parasites
We carried out a parasitological survey in the Municipal Natural Park of ‘Cancão’, located in the municipality of Serra do Navio, Amapá, Brazil (0°54′8.68″N, 52°0′19.62″W), from 2015 to 2019. We collected 28 specimens of B. boans, 27 of Boana dentei (Bokermann, 1967) and 51 of Boana multifasciata (Günther, 1859) through an active search (permission number SISBIO: no. 48102-2/IBAMA/ICMBio).
The hosts were anaesthetized and euthanized with ketamine 2%, measured, weighed and necropsied for helminth search. All internal organs were placed in Petri dishes with saline solution (NaCl 0.9%) and examined in a LEICA EZ4 stereomicroscope. The helminths found were cleaned in saline solution, killed in heated 70% ethanol and preserved in the same solution at room temperature. For morphological and morphometric analysis, the nematodes were hydrated in distilled water, cleared in Aman's lactophenol 20%, mounted on temporary slides and examined under an Olympus BX41 microscope (Olympus, Tokyo, Japan) coupled with a drawing tube (without zoom adjustment). The illustrations were prepared in CorelDraw 2018 software and processed using Adobe Photoshop Version 21.0.2 software.
The measurements are presented as the values of the holotype followed by the mean of the paratypes and range in parentheses (reported in micrometres unless otherwise indicated); the metrics of nematodes obtained from different host species are given in Table 1. The prevalence and mean intensity are according to Bush et al. (Reference Bush, Lafferty, Lotz and Shostak1997) and Reiczigel et al. (Reference Reiczigel, Marozzi, Fábián and Rózsa2019). The classification of amphibian hosts follows that of Segalla et al. (Reference Segalla, Berneck, Canedo, Caramashi, Cruz, Garcia, Grant, Haddad, Lourenço, Mângia, Mott, Nascimento, Toledo, Werneck and Langone2021) and Frost (Reference Frost2022). We deposited the type series in the Helminthological Collection of Oswaldo Cruz Institute (CHIOC), Brazil.
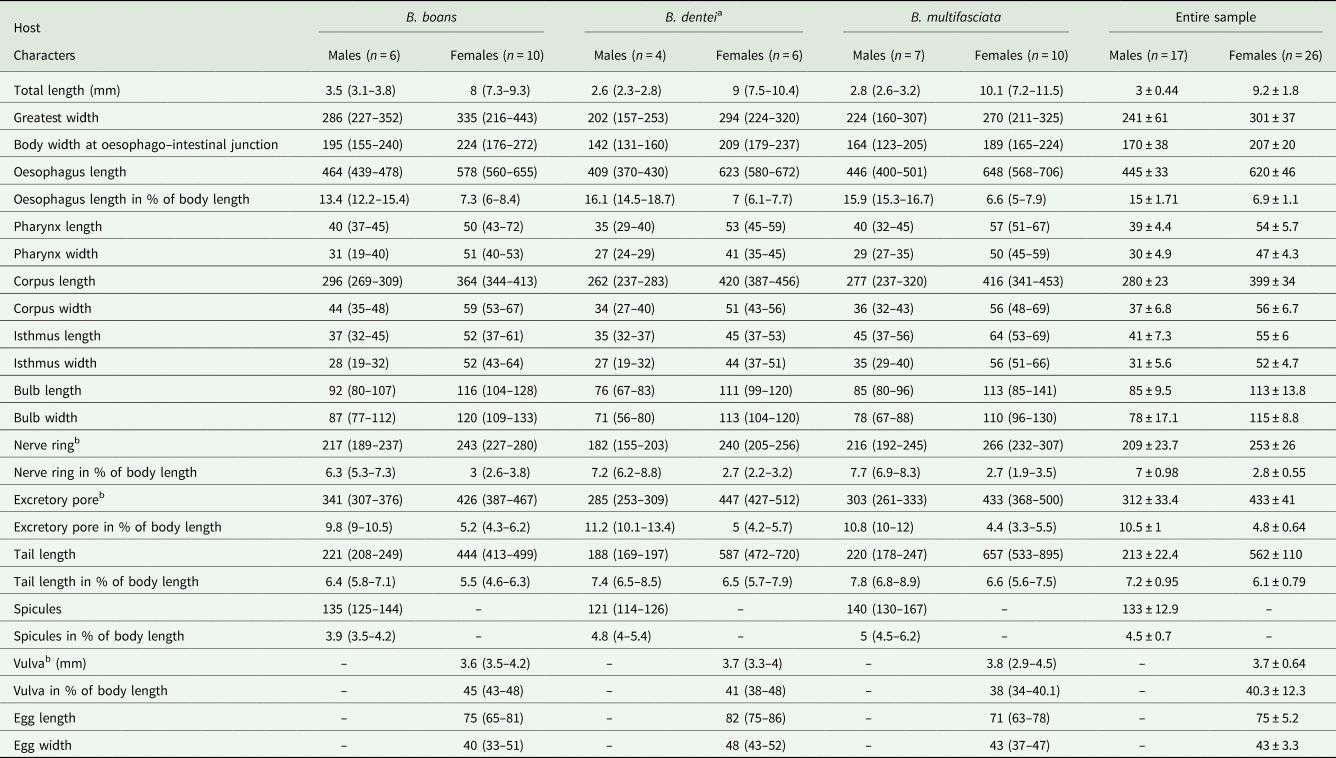
Table 1. Morphometric data of Cosmocercoides amapari n. sp. from Boana boans, Boana dentei and Boana multifasciata
All measurements are in micrometres unless otherwise indicated.
a Type series.
b From anterior end.
Some specimens were post-fixed in 1% osmium tetroxide, dehydrated in an increasing ethanol series and critical point dried in carbon dioxide. The nematodes were mounted on metallic stubs, coated with gold palladium and examined under a Vega3 (TESCAN, Brno, Czech Republic) scanning electron microscope in the Laboratory of Structural Biology, Biological Sciences Institute, Federal University of Pará (UFPA), Brazil.
Molecular analyses and phylogenetic study
For molecular analyses, 1 male specimen was transferred to a microtube with 100% ethanol and stored in a freezer at −20°C; the anterior and posterior portions were cut and analysed by light microscopy for morphological identification of the analysed sample. Genomic DNA was extracted using a Chelex Molecular Biology Grade Resin Kit, according to the manufacturer's instructions. The partial fragment of the mitochondrial cytochrome c oxidase subunit 1 (cox1) was amplified by polymerase chain reaction (PCR), using specific primers and cycles condition following the protocols established by Chen et al. (Reference Chen, Zhang, Nakao and Li2018a). The PCR products were visualized on a 1% agarose gel to determine the yield and size of the amplified fragments and were purified using a QIAquick PCR Purification kit. The amplicons’ sequence reaction followed the protocol of the Big Dye® Terminator v.3.1 Cycle Sequencing kit, and were sequenced in a DNA ABI 3730 DNA Analyzer at the Human Genome Stem Cell Research Center, Biosciences Institute, University of São Paulo (USP), Brazil.
For phylogenetic analyses, the obtained sequences were edited using Geneious 7.1.3 software (Kearse et al., Reference Kearse, Moir, Wilson, Stones-Havas, Cheung, Sturrock and Drummond2012) and compared (using BLAST algorithm) with the data deposited in the National Center for Biotechnology Information (NCBI) (htttp://www.ncbi.nml.nih.gov). The sequences were aligned and trimmed using Muscle (Edgar, Reference Edgar2004) in Geneious 7.1.3 software (Kearse et al., Reference Kearse, Moir, Wilson, Stones-Havas, Cheung, Sturrock and Drummond2012). The stop codons were verified according to the translation frame and parameter for invertebrate mitochondrial DNA (translation frame 1, invertebrate mitochondrial), using Geneious 7.1.3 (Kearse et al., Reference Kearse, Moir, Wilson, Stones-Havas, Cheung, Sturrock and Drummond2012). Regions poorly aligned and characters with gaps in any sequences were excluded from subsequent analyses (Tran et al., Reference Tran, Sato and Luc2015).
The phylogenetic trees were performed with maximum-likelihood (ML) using RAxML (Guindon and Gascuel, Reference Guindon and Gascuel2003) and the analyses were carried out in CIPRES Science Gateway (Miller et al., Reference Miller, Pfeiffer and Schwartz2010). ML inference was implemented using bootstrap support values of 1000 repetitions, and only nodes with bootstrap values greater than 70% were considered well-supported. The trees were edited using FigTree v1.3.1 software (Rambaut, Reference Rambaut2009). We used Falcaustra sp. and Falcaustra sinensis Liu et al., 2011 (Nematoda: Kathlaniidae) as an outgroup (access numbers: MN729572 and MF113223, respectively).
Results
Systematics
Family: Cosmocercidae Travassos, 1925
Genus: Cosmocercoides Wilkie, 1930
Species: Cosmocercoides amapari n. sp. Rebêlo, Santos and Melo , 2022
Taxonomic summary
Type host: Boana dentei (Bokermann, 1967) (Amphibia: Hylidae: Hylinae).
Additional hosts: Boana boans (Linnaeus, 1758) and Boana multifasciata (Günther, 1859) (Amphibia: Hylidae: Hylinae).
Type locality: Cancão Municipal Natural Park, Serra do Navio municipality, Amapá, Brazil (0°54′8.68″N, 52°0′19.62″W).
Site of infection: Large intestine.
Infection parameters: Boana dentei, prevalence 11.1% (3 infected hosts out of 27), mean intensity 5 (1–8); Boana boans, prevalence 10.71% (3 infected hosts out of 28), mean intensity 11.7 (3–18); Boana multifasciata, prevalence 19.61% (10 infected hosts out of 51), mean intensity 4.11 (1–11).
Type material: Holotype, male (CHIOC 39303a); allotype, female (CHIOC 39303b) and paratypes, 3 males (CHIOC 39303c), 5 females (CHIOC 39303d) were deposited in the Helminthological Collection of Oswaldo Cruz Institute.
Additional material: Vouchers for 13 males (CHIOC 39304–39305a) and 20 females (CHIOC 39304–39305b) were deposited in the Helminthological Collection of Oswaldo Cruz Institute.
GenBank accession number: OQ288108
ZooBank registration: urn:lsid:zoobank.org:pub:7823C8CE-89D7-4BC2-B74F-A6AAC3D54D2E
Etymology: The new species is named after the Amapari river that rises in the municipality of Serra do Navio and bathes Amapá.
General. Small and cylindrical nematodes (Fig. 1A). Cuticle with fine transverse striations. Somatic papillae present (Fig. 2A and B). Sexual dimorphism evident, females larger than males. Lateral alae present in both sexes, extending near the nerve ring region to anus in females and at the level of precloacal rosette papillae in males (Figs 1A and 3A–C). Oral opening triangular, surrounded by 3 distinct lips: dorsal lip with 2 sessile papillae, 2 subventral lips with 1 ventral sessile papilla and lateral amphidial pores (Figs 2C and 1B). Oesophagus divided into pharynx, cylindrical corpus, small isthmus and well-developed oesophageal bulb with evident valvular apparatus (Fig. 2A and B). Nerve ring situated in the middle of oesophageal corpus (Fig. 2A and B). Excretory pore anterior to isthmus (Figs 2A and B and 1C). Tail conical, sharply pointed in both sexes (Figs 2D, F–J; 1F and 3C).
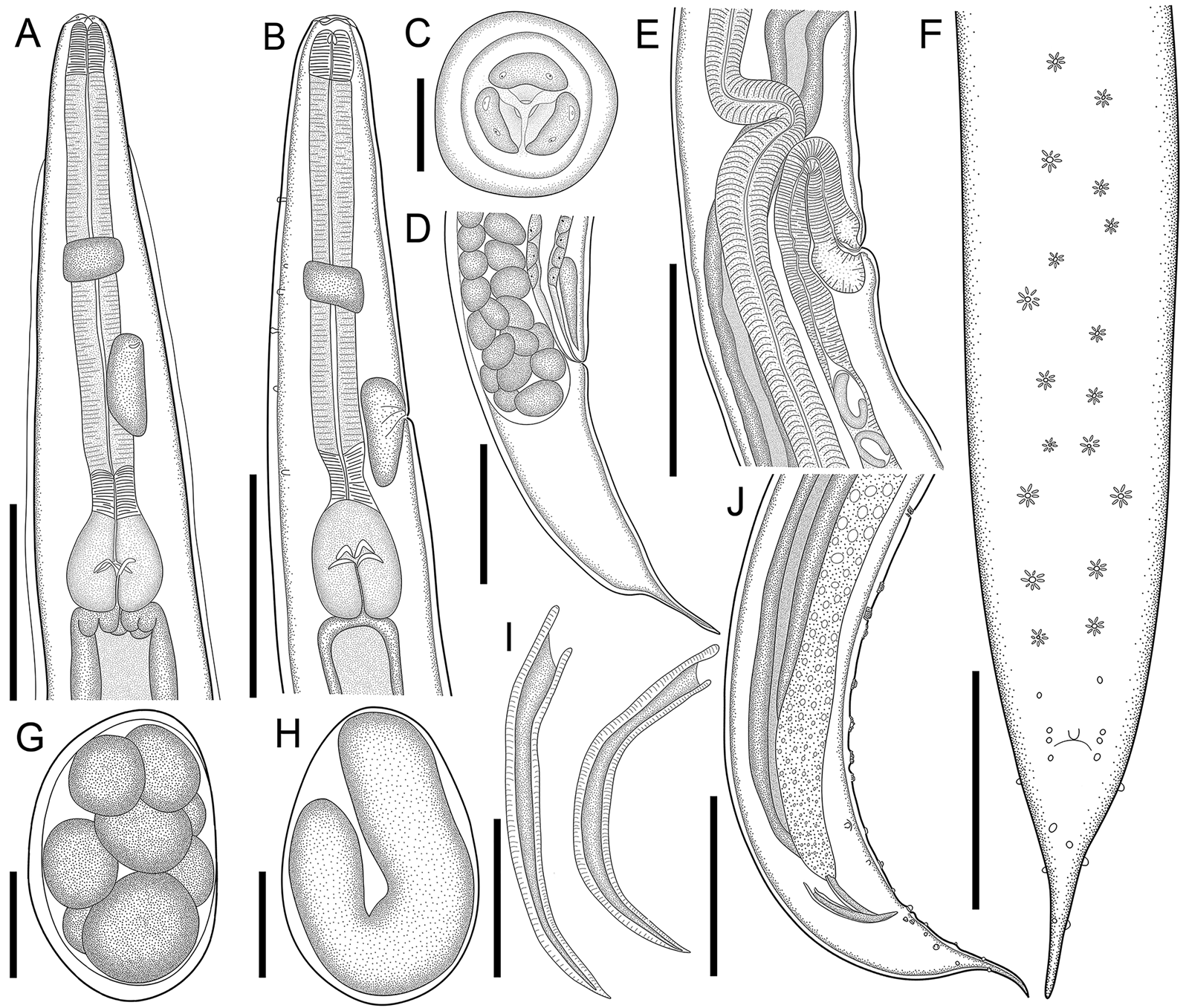
Fig. 1. Line drawings of C. amapari n. sp. from Brazilian Amazon. (A) Anterior end of male, lateral view; (B) anterior end of female, lateral view; (C) anterior end of male, apical view; (D) posterior end of female, lateral view; (E) vulva region, lateral view; (F) posterior end of male, ventral view; (G) egg in morula stage; (H) embryonated egg in the uterus; (I) spicules; (J) posterior end of male, lateral view. Scale bars: A = 150 μm; B, D, E, F, J = 200 μm; C, G, H = 25 μm; I = 100 μm.
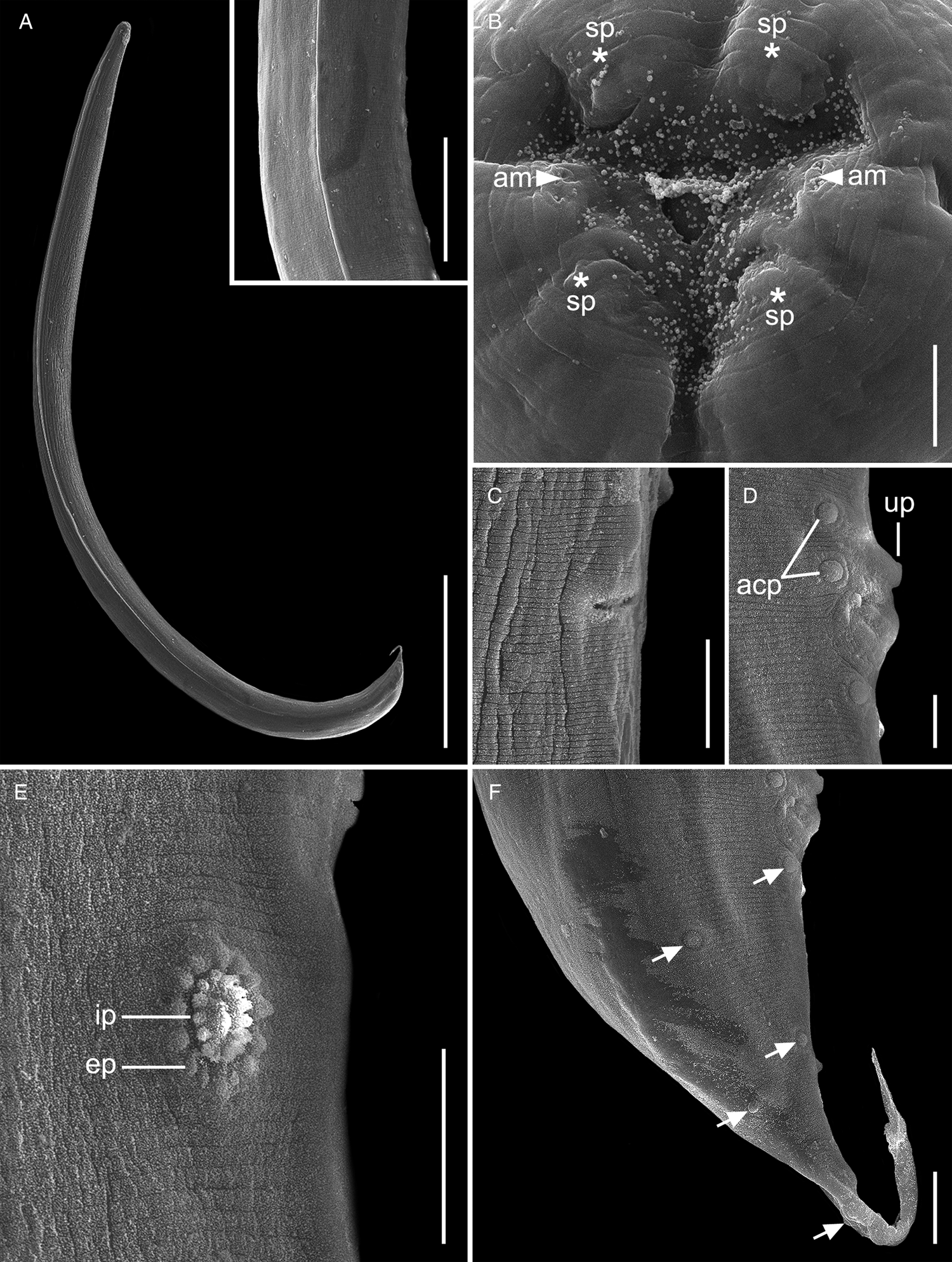
Fig. 2. Scanning electron micrographs of Cosmocercoides amapari n. sp. from Brazilian Amazon, males. (A) Entire body; inset: details of lateral alae; (B) anterior end, apical view (arrows: am – amphidial pores; asterisk: sp – papillae); (C) excretory pore; (D) disposition of adcloacal papillae (arrows: acp – adcloacal papillae; up – large unpaired papilla); (E) details of rosette papillae (line: ep – external punctations; ip – internal punctations); (F) disposition of postcloacal papillae (arrows: pcp – postcloacal papillae). Scale bars: A = 500 μm; B = 5 μm; C = 20 μm; D = 10 μm; E, F = 20 μm; inset: 100 μm.
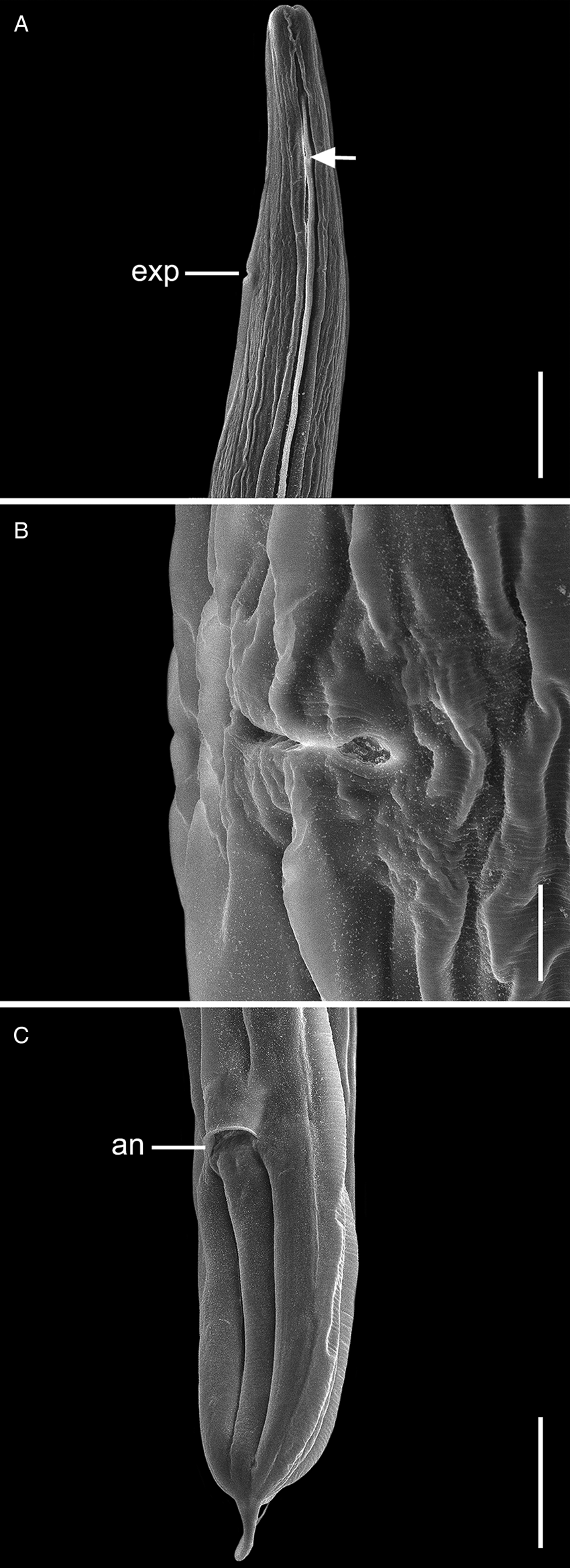
Fig. 3. Scanning electron micrographs of C. amapari n. sp. from Brazilian Amazon, females. (A) Anterior end (arrows: lateral alae; line: exp – excretory pore); (B) vulva region; (C) posterior end (line: an – anus). Scale bars: A = 100 μm; B = 20 μm; C = 100 μm.
Males (based on the holotype and 3 paratypes, all mature specimens). Total length 2.6; 2.5 (2.3–2.8) mm. Maximum width 197; 203 (157–253). Body width at oesophago–intestinal junction 136; 143 (131–160). Oesophagus 430; 402 (370–430) in total length, corresponding to 16.5; 16 (14.5–18.7)% of the body length; pharynx 35; 35 (29–40) × 24; 28 (27–29); corpus 283; 255 (237–275) × 35; 33 (27–40); isthmus 37; 35 (32–37) × 29; 27 (19–32); bulb 75; 77 (67–83) × 72; 71 (56–80). Nerve ring located at 189; 180 (155–203) from anterior extremity, corresponding to 7.3; 7.2 (6.2–8.8)% of the body length; excretory pore located at 280; 286 (253–309) from anterior extremity, corresponding to 10.8; 11.4 (10.1–13.4)% of the body length. Spicules short, equal, slightly curved ventrally, proximal ends expanded, distal ends sharply pointed 119; 122 (114–126) long, corresponding to 4.6; 4.9 (4–5.4)% of the body length (Fig. 2I). Gubernaculum absent. Caudal papillae arranged as follows: 9–10 pairs of ventral precloacal rosette papillae, distributed in 2 longitudinal rows, paired irregularly; 1 pair of precloacal simple papilla; 2 pairs of paracloacal simple papillae and 1 unpaired papilla at anterior cloacal lip; 5 pairs of postcloacal simple papillae (first and third pairs ventral, second, fourth and fifth ventrolateral) (Fig. 2F–J and 1E and F). Each rosette composed of 2 complete rings of about 13–15 punctations around central papilla (Fig. 1E). Somatic papillae present in subdorsal rows along the body. Testis single, tubular, flexing posteriorly at last third of the body length. Tail conical 169; 194 (188–197) long, corresponding to 6.5; 7.7 (6.7–8.6)% of the body length.
Females (based on the allotype and 5 paratypes, all gravid specimens). Total length 7.5; 9.3 (8.3–10.4) mm. Maximum width 317; 290 (224–320). Body width at oesophago–intestinal 237; 204 (179–232). Oesophagus 615; 630 (580–672) in total length, corresponding to 8.2; 6.8 (6.1–7.7)% of the body length; pharynx 45; 54 (51–59) × 43; 40 (35–45) long; corpus 400; 424 (387–456) × 53; 51 (43–56) long; isthmus 53; 43 (37–51) × 51; 443 (37–45) long; bulb 117; 109 (99–120) × 120; 111 (104–120). Nerve ring located at 237; 240 (205–256) from anterior extremity, corresponding to 3.2; 2.6 (2.2–3.1)% of the body length; excretory pore located at 427; 451 (432–512) from anterior extremity, corresponding to 5.7; 4.9 (4.2–5.4)% of the body length. Vulva slightly pre-equatorial 3.4; 3.8 (3.3–4) mm from anterior extremity, corresponding to 45; 41 (38–48)% of the body length (Figs 2E and 3B). Amphidelphic genital system formed by an anterior muscular vagina directed anteriorly in proximal half; flexed posteriorly in distal portion, divided into 1 anterior and a posterior uterus (Fig. 2E). Ovary directed anteriorly to vagina not extending beyond bulb. Uteri, containing numerous eggs in different stages of development, embryonated eggs and free-stage larvae close to vulva (Fig. 2G and H). Eggs size 75; 83 (82–86) × 45; 48 (43–52) (based on 10 embryonated eggs). Tail conical 595; 586 (472–720) long, corresponding to 7.9; 6.3 (5.7–7.1)% of the body length.
Variability: Values of body length and related metrical features in specimens from B. boans were somewhat higher than those of the specimens obtained from B. dentei and B. multifasciata, although most of the metric features overlapped (Table 1). The pattern of caudal papillae did not vary among the samples of the 3 hosts analysed.
Remarks
The new species belongs to the genus Cosmocercoides based on molecular data and the general morphology of males, as the presence of papillae surrounded by punctations (rosette papillae) and not ornamented with sclerotized supports (plectanes) on their caudal region. The main morphological characteristics used to distinguish species of the genus are the pattern of caudal papillae; size and shape of spicules and gubernaculum (if present), length of tail and the presence or absence of lateral alae and somatic papillae (Draghi et al., Reference Draghi, Drago and Lunaschi2020; Dos Anjos et al., Reference Dos Anjos, Oda, Campião, Ávila, Dos Santos, Dos Santos, Almeida, Melo and Rodrigues2021).
The 28 species reported for the genus Cosmocercoides can be divided into 2 groups based on the presence and absence of gubernaculum. The first group includes 23 species in which the gubernaculum is present: Cosmocercoides tibetanum Baylis, 1927; C. dukae Holl, 1928; C. pulcher Wilkie, Reference Wilkie1930; Cosmocercoides variabilis Harwood, Reference Harwood1930; Cosmocercoides skrajabini Ivanitzky, 1940; Cosmocercoides bufonis Karve, 1944; Cosmocercoides multipapillata Khera, 1958; Cosmocercoides rickae Ogden, 1966; Cosmocercoides nainitalensis Arya, 1979; Cosmocercoides barodensis Rao, 1979; Cosmocercoides lanceolatus Rao, 1979; Cosmocercoides oligodentis Wang et al., 1981; Cosmocercoides ranae Wang et al., 1981; Cosmocercoides speleomantis Ricci, 1987; Cosmocercoides tridens Hasegawa, 1989; Cosmocercoides karnatacaensis Rizvi, 2009; C. sauria Ávila et al., 2010; Cosmocercoides kiliwai Martínez-Salazar et al., 2013; Cosmocercoides himalayanus Rizvi and Bursey, 2014; Cosmocercoides malayensis Bursey et al., 2015; C. tonkinensis Tran et al., Reference Tran, Sato and Luc2015; C. qingtianensis Chen et al., Reference Chen, Zhang, Nakao and Li2018 and C. wuyiensis Liu et al., Reference Liu, Yu, Shu, Zhao, Fang and Wu2019.
Of the above-mentioned species, the number of caudal papillae in the new species is similar to the following: C. bufonis, C. ranae and C. wuyiensis. However, the new species has a pattern of rosette papillae of 18–20:0:0 and a pattern of simple papillae of 2:4:10 vs C. bufonis 18–26:2:6 and simple papillae of 0:0:20. Additionally, the new species differ by having smaller spicules compared to C. bufonis (114–126 in C. amapari n. sp. vs 190–260 in C. bufonis), and by the presence of somatic papillae (absent in C. bufonis). The new taxon can also be easily distinguished from C. ranae by the pattern of rosette papillae (18–20:0:0 in C. amapari vs 20:0:0 in C. ranae) and simple caudal papillae (2:4:10 in C. amapari vs 8:0:8 in C. ranae). Moreover, they also differ by the length of the spicules, 114–126 in the new species and 192 in C. ranae and by the presence of somatic papillae (absent in C. ranae).
Cosmocercoides wuyiensis differs from the new species by the number and pattern of rosette and simple papilla (18–20:0:0 rosette papillae and 2:4:10 simple papillae in C. wuyiensis). They also differ by length of the spicules, in C. amapari the spicules are equal in length, measuring 114–126 while in C. wuyiensis the spicules are unequal in length and width (151–163 the smallest and 189–206 the widest); additionally, the gubernaculum is absent in the new species and present in C. wuyiensis.
The new species belongs to the group of species with gubernaculum absent: Cosmocercoides microhylae Wan et al., 1978 from Paleoartic; Cosmocercoides kumaoni Arya, 1991 from Oriental region and C. lilloi Ramallo et al., Reference Ramallo, Bursey and Goldberg2007; C. latrans Draghi et al., Reference Draghi, Drago and Lunaschi2020 and C. meridionalis Anjos et al., 2021 from Neotropics.
The new species differs from C. microhylae, by the absence of somatic papillae along the body (present in C. amapari n. sp.), a different pattern of simple caudal papillae (20:2:8 in C. microhylae vs 2:4:10 in C. amapari n. sp.); the males are smaller in C. microhylae (2.2 mm total length vs 2.3–3.8 mm in C. amapari n. sp.); also differ in the length of the spicules, smaller in C. amapari n. sp. (114–126) and larger in C. microhylae (140) and by having a shorter tail (157 in C. microhylae vs 169–249 in C. amapari n. sp.).
Cosmocercoides amapari n. sp. can be distinguished from C. kumaoni (Oriental species) by the presence of a singular hook-shaped structure near the precloacal region (absent in C. amapari n. sp.); the absence of simple caudal papillae on the tail of males (present in C. amapari n. sp.), a different pattern of rosette papillae (24:2:10 in C. kumaoni vs 18–20:0:0 in C. amapari n. sp.) and shorter tail (130–150 in C. kumaoni vs 169–249 in C. amapari n. sp.).
Among Neotropical species, the new species differs from C. lilloi by the presence of postcloacal rosette papillae (absent in C. amapari n. sp.); and the absence of simple caudal papillae, unpaired papilla at the anterior cloacal lip, lateral alae and somatic papillae (all these morphological traits were observed in C. amapari n. sp.). When compared to C. latrans, C. amapari n. sp. presents a higher number of precloacal rosette papillae (6–8 in C. latrans vs 18–20 in C. amapari n. sp.) and simple caudal papillae (0:2:8 in C. latrans vs 2:4:10 in C. amapari n. sp.).
The new species resembles C. meridionalis in some metric characters, such as body size, the distance of the nerve ring from the anterior end and the spicules length. However, C. meridionalis has a longer oesophagus (520–650 in C. meridionalis vs 370–501 in C. amapari n. sp.), different position of the excretory pore (426–571 in C. meridionalis vs 253–376 in C. amapari n. sp. from anterior extremity) and longer tail (290–446 in C. meridionalis vs 169–249 in C. amapari n. sp.). In addition, C. meridionalis has different pattern and distribution of rosette papillae (22:2:2 in C. meridionalis vs 18–20:0:0 in C. amapari n. sp.), with paracloacal and postcloacal rosette papillae (absent in C. amapari n. sp.) and a small number of simple caudal papillae on the tail (0:0:6 in C. meridionalis vs 2:4:10 in C. amapari n. sp.).
Cosmocercoides sauria can be easily distinguished from C. amapari by the presence of gubernaculum in former species. Additionally, C. sauria also differs from C. amapari n. sp. in body length (1.3 in C. sauria vs 2.3–3.8 in C. amapari n. sp.), by having shorter spicules (104 in C. sauria vs 114–167 in C. amapari n. sp.), shorter tail (54 in C. sauria vs 169–249 in C. amapari n. sp.), absence of somatic papillae (present in C. amapari n. sp.), smaller number of precloacal rosette papillae (8 in C. sauria vs 18–20 in C. amapari n. sp.) and simple caudal papillae (0:1:4 in C. sauria vs 2:4:10 in C. amapari n. sp.).
Molecular analyses and phylogenetic study
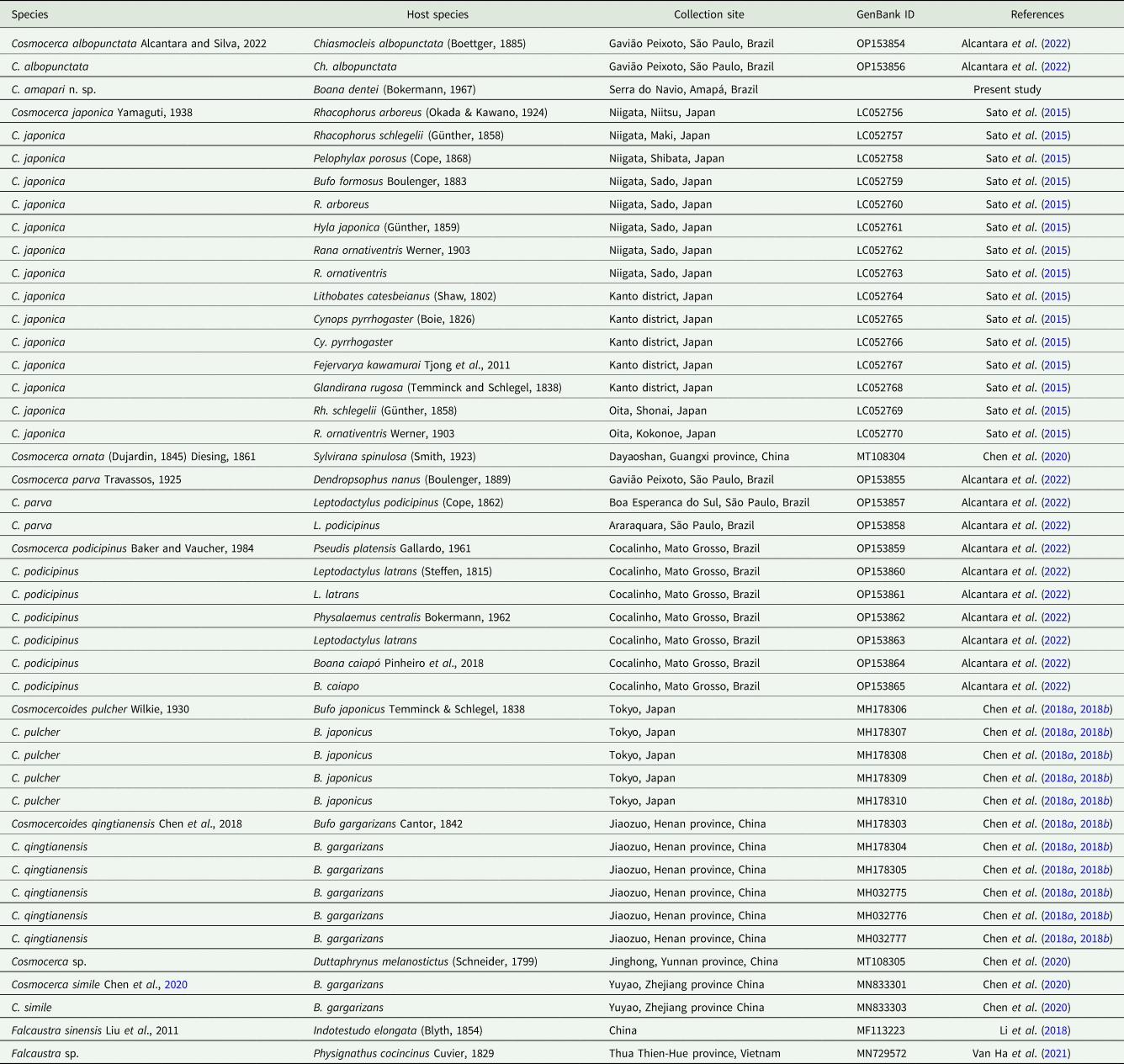
The mitochondrial cox1 sequence obtained from C. amapari n. sp. has 417 bp in length. Our search for similar sequences deposited in GenBank resulted in 3 other cox1 sequences from Cosmocercoides (C. pulcher – accession no. MH178310, C. qingtianensis – accession no. MH178305 and C. wuyiensis accession no. MK956953) (Table 2). Pairwise comparison between C. amapari n. sp. and C. pulcher showed 19% nucleotide divergence, and the divergence between C. amapari n. sp. and C. qingtianensis was 19% (supplementary Table 1). We did not include C. wuyiensis in our analysis and comparisons due to the number of indels in the sequences.
Table 2. Representatives of Cosmocercoides spp. used for phylogenetic analyses related to information on host, locality and GenBank ID
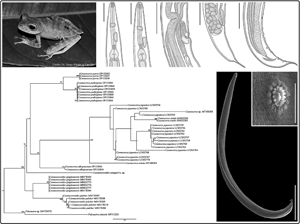
ML phylogenetic analysis revealed that the new species sequence was related as a sister group of a large clade with all the Cosmocerca spp. (70% bootstrap). Sequences of C. pulcher and C. qingtianensis parasites of Bufo japonicus formosus Matsui, 1984 from China and Bufo gargarizans Cantor, 1842 from Tokyo, Japan, respectively, formed a monophyletic cluster (40% bootstrap) sister to the large clade that clustered sequences from Cosmocerca spp. + C. amapari. The clade that groups the Cosmocerca spp. sequences were subdivided into 2; a smaller clade that groups together sequences from Cosmocerca parva parasite of Dendropsophus nanus (Boulenger, 1889) and Leptodactylus podicipinus (Cope, 1862), both from Brazil and Cosmocerca podicipinus (94% bootstrap) parasite of Pseudis platensis Gallardo, 1961, Leptodactylus luctator (Steffen, 1815), Physalaemus centralis Bokermann, 1962, Boana caiapo Pinheiro et al., 2018 and a larger clade that is subdivided into a monophyletic group formed by sequences from Cosmocerca albopunctata (94% bootstrap) parasite of Chiasmocleis albopunctata (Boettger, 1885) from Brazil and a branch with a sequence from Cosmocerca ornata parasite of Sylvirana spinulosa (Smith, 1923) that is related as sister species of the clade with sequences from Cosmocerca japonica + Cosmocerca simile (100% bootstrap) parasites of multiple hosts (for C. japonica) from Japan and C. simile parasite of Bufo gargarizans from China (Fig. 4).

Fig. 4. ML phylogenetic topology based on the partial cox1 sequence data using Falcaustra sp. and Falcaustra sinensis Liu et al., 2011 (Nematoda: Kathlaniidae) as outgroup indicating the position of C. amapari n. sp. and the phylogenetic relationships of the representatives of the Cosmocercidae. GenBank accession numbers follow each taxon. Support values are above or below nodes: bootstrap scores <70% are not shown or are represented by a dash. Branch-length scale bar indicates the number of substitutions per site.
Discussion
In our study, the new species showed variability of metrical features in the samples of the different hosts analysed. Similar results were also reported for C. pulcher and C. variabilis, where the authors found those species parasitizing a broad spectrum of hosts (Harwood, Reference Harwood1930; Wilkie, Reference Wilkie1930; Vanderburgh and Anderson, Reference Vanderburgh and Anderson1987; Joy and Bunten, Reference Joy and Bunten1997; Bursey et al., Reference Bursey, Goldberg and Telford2007; Bursey and Brooks, Reference Bursey and Brooks2011; Tran et al., Reference Tran, Sato and Luc2015; Chen et al., Reference Chen, Zhang, Nakao and Li2018a).
The monophyly of the Cosmocercidae family has been corroborated by several studies (Tran et al., Reference Tran, Sato and Luc2015; Sinsch et al., Reference Sinsch, Heneberg, Těšínský, Balczun and Scheid2019; Sinsch et al., Reference Sinsch, Dehling, Scheid and Balczun2020; Ni et al., Reference Ni, Chen, Xu, Gu and Li2022); nonetheless, due to the low sampling of taxa, geographic limitations and few molecular data available, the phylogenetic relationships between the genera of the family are still unclear. Sinsch et al. (Reference Sinsch, Dehling, Scheid and Balczun2020) and Chen et al. (Reference Chen, Zhang, Feng and Li2020) during phylogenetic studies, using the 18S and internal transcribed spacer 1 ribosomal markers, demonstrated that the genera Cosmocerca and Cosmocercoides are phylogenetically close related. However, recent studies also showed that Cosmocerca and Aplectana are phylogenetically closer (Chen et al., Reference Chen, Gu, Ni and Li2021; Harnoster et al., Reference Harnoster, Du Preez and Svitin2022). Additionally, the species Cosmocerca longicauda (Linstow, 1885), which in the analyses of Chen et al. (Reference Chen, Zhang and Li2018b), Sinsch et al. (Reference Sinsch, Heneberg, Těšínský, Balczun and Scheid2019) and Sinsch et al. (Reference Sinsch, Dehling, Scheid and Balczun2020) are closely related to Cosmocercoides (based on 18S region), was not included in these works. These results reinforce that the interspecific relationships of Cosmocercidae still need to cover more species of the family.
In our analyses, after adding sequence from the new taxon, we found the same results as previous authors. Our phylogeny showed a clade formed by C. pulcher + C. qingtianensis (with low support). In phylogenetic studies using 18S and 28S from the Oriental region, the authors recovered a clade formed by sequences of the species C. pulcher + C. tonkinensis and C. qingtianensis or C. pulcher and C. tonkinensis (Tran et al., Reference Tran, Sato and Luc2015; Chen et al., Reference Chen, Zhang, Feng and Li2020, Reference Chen, Gu, Ni and Li2021; Harnoster et al., Reference Harnoster, Du Preez and Svitin2022; Ni et al., Reference Ni, Chen, Xu, Gu and Li2022).
Alcantara et al. (Reference Alcantara, Ebert, Müller, Úngari, Ferreira-Silva, Emmerich, Santos, O'Dwyer and da Silva2022) when describing Cosmocerca albopunctata Alcantara and Silva, 2022, and including sequences from Cosmocerca spp. from the Neotropics based on the molecular marker cox1, also recovered Cosmocercoides as a monophyletic group, but with low support. However, our analysis showed that C. amapari n. sp. was grouped with species of Cosmocerca (with low support), indicating that its phylogenetic position within the group can change with the addition of new sequences from the same region or from species that have not yet been discovered.
Thus, the low support values show that the small amount of available sequences and from different biogeographic regions are factors that may be limiting the analysis. Therefore, to confirm the monophyly of the genus, it will be necessary to add new sequences from other species of Cosmocercoides. Additionally, the present work and the work of Alcantara et al. (Reference Alcantara, Ebert, Müller, Úngari, Ferreira-Silva, Emmerich, Santos, O'Dwyer and da Silva2022) are the only studies that analysed the phylogeny of Cosmocercidae using the cox1 marker. Thus, based on our findings and those from recent studies (Chen et al., Reference Chen, Gu, Ni and Li2021; Alcantara et al., Reference Alcantara, Ebert, Müller, Úngari, Ferreira-Silva, Emmerich, Santos, O'Dwyer and da Silva2022) we raised the hypothesis that Cosmocercoides spp. might have an independent diversification process in the Neotropics.
The interspecific nucleotide divergence observed in cox1 mtDNA between C. amapari n. sp. and C. qingtianensis was 19% and that for C. amapari n. sp. vs C. pulcher varied from 19 to 23%, supporting the genetic differences between the new taxon and these species. The divergence values among species from Oriental realm were even lower (varied from 12.50 to 18.24%); however, it presents range values commonly found for Cosmocercidae (see Chen et al., Reference Chen, Zhang, Nakao and Li2018a; Alcantara et al., Reference Alcantara, Ebert, Müller, Úngari, Ferreira-Silva, Emmerich, Santos, O'Dwyer and da Silva2022). The high values of divergence found here may reflect the geographical distance and/or distinct morphological features, especially the gubernaculum (present in C. qingtianensis and C. pulcher, and absent in the new taxon). According to Chen et al. (Reference Chen, Zhang, Nakao and Li2018a) and Liu et al. (Reference Liu, Yu, Shu, Zhao, Fang and Wu2019), the molecular marker cox1 is the most suitable, practical, rapid and accurate for identifying and differentiating Cosmocercoides species; these authors observed pieces of evidence of interspecific variation of nucleotides in this region, even when the analysed species had morphological similarity and belonged to the same biogeographic area.
Our phylogenetic analyses also indicate that C. pulcher and C. podicipinus may represent species complex; however, additional analysis should be carried out to give support to this hypothesis. The sequences of C. japonica are separated into 4 small clades, not well supported, which reinforces the need for taxonomic review and evaluation of the sequences deposited in the database as suggested by Alcantara et al. (Reference Alcantara, Ebert, Müller, Úngari, Ferreira-Silva, Emmerich, Santos, O'Dwyer and da Silva2022).
Finally, morphological and molecular evidence based on the cox1 region revealed that the nematode collected in B. boans, B. dentei and B. multifasciata represent a new species of Cosmocercoides. Our study also presents important results, as it is the first species of Cosmocercoides in Brazil and in the Neotropical region with genetic data, adds more information to Cosmocercidae and reinforces the need for further morphological and molecular studies that clarify family evolutionary relationships.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182022001767.
Acknowledgements
We are grateful to Profa. Cynthya Elizabeth González from the Centro de Ecología Aplicada del Litoral (CECOAL), Universidad Nacional del Nordeste, Corrientes, Argentina for her contributions to this work. We appreciate the help of Profa. Edilene Oliveira da Silva and MSc.Yuri Wilkens from the Federal University of Pará, Belém, Brazil with the SEM analysis; we are grateful to students from the Laboratory of Cellular Biology and Helminthology ‘Profa. Dra. Reinalda Marisa Lanfredi’ (Federal University of Pará, Belém, Brazil); and also to students from the Laboratory of Herpetology of the Federal University of Amapá (Federal University of Amapá, Macapá, Brazil); in addition, we thank professionals from the Chico Mendes Institute of Biodiversity Conservation for providing us permission to collect specimens and to PROPESP/UFPA and Prof. Dr Edson A. Adriano from the Laboratory of Ecology and Evolution, Federal University of São Paulo (UNIFESP) for their support in carrying out molecular analyses.
Author's contributions
G. L. Rebêlo wrote the main draft and prepared images. A. N. Santos, R. F. Jesus and C. E. Costa-Campos helped with specimen observations and SEM analysis, reviewing and writing the manuscript. A. N. Santos and M. I. Müller carried out PCR and sequencing. L. F. S. Tavares-Costa, M. R. Dias-Souza and M. I. Müller helped with phylogeny. J. N. dos Santos and F. T. V. Melo collected the specimens, helped with morphological and molecular analysis, wrote the manuscript, revised and prepared the line drawings. All authors reviewed the manuscript.
Financial support
This work was supported by CAPES/UFPA and the National Council for Scientific and Technological Development (CNPq) (grant number 431809/2018-6 Universal); Productivity Scholarship Grant (CNPq) to J. N. dos Santos (process no. 305552/2019-8); to F. T. V. Melo (CNPq) (process: 304955/2018-3). M. I. Müller was supported by a postdoctoral scholarship from the São Paulo Research Foundation (FAPESP) (grant no. 2017/16546-3) (FAPESPA/CNPQ PRONEM 01/2021, process no. 794027/2013). This study is part of the M.Sc. dissertation of G. L. Rebêlo, as part of the Postgraduate Program in Biology of Infectious and Parasitic Agents (BAIP-ICB-UFPA).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
All applicable institutional, national and international guidelines for the care and use of animals were followed. Host specimens were collected under permits from Institute for the Environment and Renewable Resources – IBAMA/ICMBio (SISBIO: no. 48102-2) and Ethics Committee on the Use of Animals of the Federal University of Para (CEUA/UFPA: no. 8341260821).