Introduction
Fish are commonly infected by a diversity of parasites, some of which appear to have subtle or undetectable effects on their hosts (Moles and Heifetz, Reference Moles and Heifetz1998; Goater et al., Reference Goater, Goater and Esch2014), whereas others can cause conspicuous host pathology, potentially impacting entire populations and communities (Lafferty, Reference Lafferty2008; Heins et al., Reference Heins, Birden and Baker2010; Goater et al., Reference Goater, Goater and Esch2014). The selective pressures imposed by parasites on hosts and responses of hosts thereto can result in host specificity, here considered to represent a parasite infecting 1 host species. Although processes of parasitism have received considerable attention in ecological research, the diversity and range of host species remains unclear (Wells and Clark, Reference Wells and Clark2019; Shim et al., Reference Shim, Peterson and Bolnick2023), especially under conditions allowing for unrestricted transmission of parasites among coincident hosts within a local community (e.g. Blasco-Costa et al., Reference Blasco-Costa, Balbuena, Raga, Kostadinova and Olson2010; McNamara et al., Reference McNamara, Mille and Cribb2014). For example, further investigation might demonstrate that assemblages of sympatric hosts are more frequently infected by phenotypically similar but evolutionarily distinct parasites than is currently known (Choudhury and Scholz, Reference Choudhury and Scholz2020). If so, the diversity of parasites might be underestimated and the structure and function of resident communities mischaracterized.
Research on lineages of tapeworms in the genus Schistocephalus (Cestoda: Diphyllobothriidea) may help us to reveal the ecological and evolutionary underpinnings of parasite diversity. These cestode parasites are trophically transmitted with complex life cycles, which is well illustrated by the life cycle of Schistocephalus solidus (Smyth, Reference Smyth1962): a free-living, planktonic coracidium larva; followed in turn by a procercoid infecting any of several cyclopoid copepods (first-intermediate host); a plerocercoid infecting a threespine stickleback (Gasterosteus aculeatus, second-intermediate host) and an adult worm reproducing in any of about 40 species of piscivorous birds (definitive host). The stickleback fish is the only obligate host in the life cycle. Almost all growth of S. solidus, from microscopic larva to macroscopic plerocercoid, required for reproduction in the definitive host occurs in the intermediate host fish, which can significantly reduce host energy reserves (Walkey and Meakins, Reference Walkey and Meakins1970; Lester, Reference Lester1971; Schultz et al., Reference Schultz, Topper and Heins2006).
Unlike other stages of the Schistocephalus life cycle, plerocercoids appear to exhibit strict specificity for particular hosts, notwithstanding ecological conditions one might expect would allow widespread transmission among co-occurring fish species. Notably, research on the first 2 species of Schistocephalus demonstrated to be biological species, S. solidus and Schistocephalus pungitii, indicates that S. solidus infects the threespine stickleback, whereas S. pungitii infects the ninespine stickleback (Pungitius pungitius) (Nishimura et al., Reference Nishimura, Heins, Andersen, Barber and Cresko2011). Early morphological and cross-infection studies (Dubinina, Reference Dubinina1959; Braten, Reference Braten1966) provided evidence of host specificity, an inference later supported by phylogenetic analyses showing that distinct lineages of Schistocephalus cestodes infect threespine and ninespine stickleback hosts, respectively, from western North America and western Europe (Nishimura et al., Reference Nishimura, Heins, Andersen, Barber and Cresko2011).
The number of fishes discovered to be intermediate hosts of Schistocephalus plerocercoids now includes freshwater sculpins (family Cottidae) from widely separated locations, including bullhead, Cottus gobio, in an Arctic river in Finland (Chubb et al., Reference Chubb, Seppälä, Luscher, Milinski and Valtonen2006); slimy sculpin, Cottus cognatus, in lakes of the Arctic region of Alaska, USA (V.B. Holland, unpublished MSc thesis, University of North Carolina at Greensboro, 2006), a lake of southwest Alaska (Harmon et al., Reference Harmon, Hilborn and Quinn2015), Lake Michigan, USA (French and Muzzall, Reference French and Muzzall2008) and the Athabasca River drainage, Alberta, Canada (Braicovich et al., Reference Braicovich, McMaster, Glozier and Marcogliese2020) and coastrange sculpin, Cottus aleuticus, in a lake of southwest Alaska (Harmon et al., Reference Harmon, Hilborn and Quinn2015). Thus, multiple species of fishes are potentially susceptible to infection by Schistocephalus cestodes, including sticklebacks and sculpins that often co-occur in lake habitats (McPhail and Lindsey, Reference McPhail and Lindsey1970). Whether host specificity extends to all or some subset of co-occurring species within local communities of sculpins and sticklebacks is unclear, as is the number of Schistocephalus species that may have diversified among fish hosts. Beyond the phylogenetic analyses of Nishimura et al. (Reference Nishimura, Heins, Andersen, Barber and Cresko2011), the only other investigation of host specificity and differentiation in Schistocephalus was completed by Chubb et al. (Reference Chubb, Seppälä, Luscher, Milinski and Valtonen2006), who named the cestode Schistocephalus cotti as a new species based on morphological and genetic differences between parasites from C. gobio and G. aculeatus. One might thus expect Schistocephalus plerocercoids of other fish hosts to exhibit morphological and genetic differences indicative of host specificity.
We examined Schistocephalus plerocercoids from co-occurring slimy sculpin, coastrange sculpin, threespine stickleback and ninespine stickleback to investigate the nature of host specificity and differentiation among fish hosts in local communities. We drew inferences based on morphological, genetic and phylogenetic comparisons of parasites from all 4 fish species sampled from 2 lakes in different river drainages in southwest Alaska. This effort builds on prior investigations of the ecology (Quinn et al., Reference Quinn, Kendall, Rich and Chasco2012) and genetics (Sprehn et al., Reference Sprehn, Blum, Quinn and Heins2015) of S. solidus in threespine stickleback from Bristol Bay (southwest Alaska, USA) that led to detection of cryptic plerocercoids in slimy sculpin and coastrange sculpin from Iliamna Lake (Harmon et al., Reference Harmon, Hilborn and Quinn2015). Initial examinations revealed that the cestodes in the 2 sculpin species exhibit more segments than those in threespine sticklebacks, consistent with the pattern reported for cestodes from cottids in Finland by Chubb et al. (Reference Chubb, Seppälä, Luscher, Milinski and Valtonen2006). Accordingly, we tested the hypothesis that the cestodes infecting sculpin and stickleback hosts correspond to 2 distinct evolutionary lineages. Given prior research illustrating that different species of stickleback hosts carry different species of Schistocephalus parasites, we also tested for finer-scale differentiation between sculpin parasites reflecting host specificity sufficient to warrant recognition of distinct species.
Materials and methods
Study sites and focal species
Lakes Aleknagik (59.7445 N, 154.1427 W) and Iliamna (59.3435 N, 154.7802 W) are part of the Wood River and Kvichak River watersheds, respectively, both of which drain into Bristol Bay, Alaska. Lake Aleknagik is smaller (83 km2 in surface area, 3.6 km3 in volume, with mean and maximum depths of 43 and 110 m) than Iliamna Lake (2622 km2 in area, 115.3 km3 in volume, with mean and maximum depths of 44 and 301 m; Burgner et al., Reference Burgner, DiCostanzo, Ellis, Harry, Hartman, Kerns, Mathisen and Royce1969). Both lakes are oligotrophic but primary and secondary production levels are higher in Aleknagik than Iliamna (Burgner et al., Reference Burgner, DiCostanzo, Ellis, Harry, Hartman, Kerns, Mathisen and Royce1969). The zooplankton communities are similar (primarily cyclopoid and calanoid copepods and cladocerans) but Aleknagik has a higher proportion of Daphnia than does Iliamna, where Bosmina is the dominant cladoceran (Hoag, Reference Hoag1972; Carter and Schindler, Reference Carter and Schindler2012; T. P. Quinn, unpublished data, 2024). In boreal freshwater ecosystems, ninespine and threespine sticklebacks and slimy and coastrange sculpins frequently co-occur (McPhail and Lindsey, Reference McPhail and Lindsey1970).
Sample collection
Threespine and ninespine stickleback were sampled from multiple locations in the limnetic and littoral zones whereas coastrange and slimy sculpins were sampled from littoral zone sites in both lakes in August and September of 2012–2015 and 2017–2019. Limnetic sampling was conducted with a towed surface net at a series of long-term monitoring sites in open water (see Arostegui et al., Reference Arostegui, Hovel and Quinn2018 for details). Littoral sampling was conducted with a hand net, beach seine or baited traps along mainland or island shorelines. Specimens were euthanized after capture with an overdose of buffered MS-222 and dissected for removal and evaluation of all Schistocephalus parasites, which were found in the body cavities. Sculpin species were identified with a dissecting microscope by the number of chin pores present: 1 – coastrange, 2 – slimy (Morrow, Reference Morrow1980). Due to wide variation in size among parasites found in fish hosts, segments were only counted (under a dissecting microscope) for specimens large enough to permit an accurate count. Parasite specimens and fish hosts were preserved in 70% ethanol and stored at room temperature.
Meristic analysis
To determine whether there was meristic evidence of parasite host specificity and differentiation (Chubb et al., Reference Chubb, Seppälä, Luscher, Milinski and Valtonen2006), parasite segment counts were compared according to host fish species using a generalized least squares (GLS) regression model to account for unequal sample sizes of Schistocephalus parasites from slimy sculpin, threespine stickleback and ninespine stickleback in both lakes, and from coastrange sculpin in Iliamna Lake (Table 1). The absence of Schistocephalus parasites in coastrange sculpin sampled from Aleknagik Lake also precluded formal testing for a host–lake interaction effect on segment counts in the model. Thus, a combined factor of host/lake (e.g. Iliamna slimy sculpin, Aleknagik slimy sculpin) was tested to account for potential between-lake variation within host species when comparing segment counts among host species. To identify the best-fit GLS model, variance structures were first compared for host, lake and host/lake in models with host/lake as a main effect. Backward selection was then conducted on the main effect following Zuur et al. (Reference Zuur, Ieno, Walker, Saveliev and Smith2009). Model selection (including identification of the optimal variance structure) was conducted with Akaike's information criterion (AIC – Akaike, Reference Akaike1974) of maximum-likelihood estimates. The identified best-fit model was then re-estimated with restricted maximum likelihood. Pairwise comparisons among host/lake combinations were conducted with Tukey multiple comparison tests using a Benjamini–Hochberg correction. Models were built and validated in R version 3.6.3 using the following packages: ‘stats’ (R Core Team, 2020), ‘nlme’ (Pinheiro et al., Reference Pinheiro, Bates, DebRoy, Sarkar, Heisterkamp, Van Willigen and Ranke2016), ‘piecewiseSEM’ (Lefcheck et al., Reference Lefcheck, Byrnes and Grace2018) and ‘multcomp’ (Hothorn et al., Reference Hothorn, Bretz and Westfall2008).
Table 1. Summary metrics of Schistocephalus parasite segment counts in different fish species from Iliamna Lake and Lake Aleknagik, Alaska

Sample size is the number of parasites examined; mean (s.d.) and range refer to the number of segments per parasite.
Genetic sequencing and analysis
To quantify genetic variation and potential differentiation of Schistocephalus plerocercoids across host species, genomic DNA was first extracted from 77 parasite specimens (20 from slimy sculpin, 33 from coastrange sculpin, 20 from threespine stickleback and 4 from ninespine stickleback), using the Qiagen DNeasy Blood and Tissue Kit according to the user manual for tissue extraction. DNA concentrations were quantified using a Nanodrop Spectrophotometer and then standardized to 20 ng μl−1. Polymerase chain reactions (PCRs) using GoTaq polymerase were performed to amplify a ~1100 bp portion of the NADH1 mitochondrial gene using primers from Nishimura et al. (Reference Nishimura, Heins, Andersen, Barber and Cresko2011) (forward: NAD 9F1 – GGGTTTGCGTCTCGGAGATGGTG; reverse: NAD 3R1 – GCGTAATCGTTGGTGGAAC). PCR amplifications involved an initial cycle of denaturation of 94°C for 3 min, 35 subsequent cycles of denaturation at 94°C for 1 min, annealing at an optimized temperature of 56°C for 1 min and extension at 72°C for 1 min, followed by a final extension step of 72°C for 10 min. Post-PCR products were cleaned using ExoSap (Thermo Fisher Scientific, Waltham, MA, USA). The resulting cleaned-PCR products were cycle-sequenced with each primer used for PCR amplification. Sanger electrophoresis was conducted on an ABI 3100xl. Sequences were cleaned and trimmed using Sequencher v5.1 (Gene Codes Corporation, Ann Arbor, MI, USA). All subsequent analyses focused on a 396 bp section that excluded low-quality and non-overlapping forward and reverse sequences of the target region. The haplotype of each parasite specimen was then determined according to nucleotide sequence variation. Nucleotide sequences representative of each unique haplotype were subsequently deposited in the GenBank database (accession numbers OR902521–OR902597).
Estimates of genetic variation and differentiation were determined according to nucleotide sequence variation. First, haplotype sequences were run through NCBI Blastn (Altschul et al., Reference Altschul, Gish, Miller, Myers and Lipman1990) to scan for homologous nucleotide sequences. Haplotype sequences also were run through Blastx for translated amino acid homology. Sequence divergence, haplotype diversity (h), number of segregating sites (S) and nucleotide diversity (π) were estimated in DnaSP 6.12.03 (Rozas et al., Reference Rozas, Ferrer-Mata, Sánchez-DelBarrio, Guirao-Rico, Librado, Ramos-Onsins and Sánchez-Gracia2017). Phylogenetic analyses were conducted on an alignment of the newly generated sequences and GenBank repository sequences of the NADH subunit ND1 gene from S. solidus, S. pungitii, S. cotti and Spirometra erinaceieuropaei (outgroup). All sequences were aligned with Clustal Omega (Goujon et al., Reference Goujon, McWilliam, Li, Valentin, Squizzato, Paern and Lopez2010) as implemented in Sequencher v. 5.1. Bayesian analysis of the alignment was performed with MrBayes 3.2.7a (Ronquist et al., Reference Ronquist, Teslenko, van der Mark, Ayres, Darling, Höhna, Larget, Liu, Suchard and Huelsenbeck2012) using a general time-reversible model with a portion of invariable sites and gamma-shaped distribution of rates across site models (GTR + I + Γ) and 2 simultaneous Markov chain Monte Carlo analyses with 4 chains for 3 × 106 generations. Trees were sampled for every 1000 generations, with a 25% burn-in and stop rule once convergence was established with the final deviation of split frequencies fell below 0.01.
Results
Meristic comparison
Overall, Schistocephalus parasites from the 2 stickleback species (n = 135) had fewer segments than the parasites in the 2 sculpin species (n = 140) (Table 1); 92.6% of the cestodes in sticklebacks had <100 segments and 85% of those in sculpins had >100 segments (Fig. 1). Regression analysis of parasite segment counts indicated a main effect of host/lake (F = 60.6, P < 0.0001) and a variance structure for host in the GLS model with the lowest AIC score and highest AIC weight (Table 2). The best-fit model (pseudo-R 2 = 0.56) identified large, significant differences in the mean segment counts between the 2 stickleback species and the 2 sculpin species both between and within lakes, except between slimy sculpin and threespine stickleback in Aleknagik Lake (Table 3). There were smaller but significant differences between lakes in segment counts of parasites from each stickleback species (e.g. threespine stickleback from Iliamna and Aleknagik), and between the stickleback species (threespine and ninespine), both within and between lakes (Table 3). In contrast, small, but significant, differences in parasite segment counts between coastrange and slimy sculpin only occurred between lakes (Table 3). That is, segment counts did not significantly differ between the cestodes in the 2 sculpin species within the lake (Iliamna) where such a comparison was possible (the absence of cestodes in coastrange sculpin sampled from Aleknagik Lake precluded comparison to those in slimy sculpin within that lake).

Figure 1. Number of segments per Schistocephalus parasite by host fish species. Within panels, the lake-specific data are presented as colour-coded, overlapping distributions (lighter shade – Iliamna; darker shade – Aleknagik; intermediate shade – overlap) with their corresponding probability density functions. Coastrange sculpins with parasites were only collected at Iliamna Lake.
Table 2. GLS model selection results for parasite segment counts, including the difference in AIC relative to the model with the lowest score (ΔAIC) and the AIC weight (AICw)

Rows above the dashed line describe the optimal variance structure, whereas rows below describe the subsequent main effect selection in models with the optimal variance structure.
Table 3. Pairwise, model-predicted differences in segment counts among host/lake combinations

Host: 3-sp, threespine stickleback; 9-sp, ninespine stickleback; Cr Sc, coastrange sculpin; Sl Sc, slimy sculpin. Lake: A, Lake Aleknagik; I, Iliamna Lake.
The mean difference (95% confidence interval) of each comparison is rounded to the nearest integer, and is calculated as the difference between the corresponding host/lake of that row minus the host/lake of that column (e.g. Aleknagik threespine stickleback, on average, exhibit 9 less segments than Aleknagik slimy sculpin). The comparison type is colour coded: within a species and between lakes – yellow, among species and within a lake – grey, among species and lakes – white.
Comparison P value: <0.05 (*), <0.01 (**), <0.001 (***).
Genetic variation and phylogenetic divergence
Parasites from sculpin hosts (accession numbers OR902521–OR902573) had 23 haplotypes with a haplotype diversity of 0.94, 28 segregating sites and a nucleotide diversity of 0.007. NCBI Blast analysis recovered 89.25% sequence similarity to S. cotti (accession numbers KT326912.1 and KT326911.1). Eighteen haplotypes among the parasites from stickleback hosts exhibited a haplotype diversity of 0.96, 60 segregating sites and a nucleotide diversity of 0.02. NCBI Blast analysis recovered a 95% similarity between parasites from threespine sticklebacks (accession numbers OR902574–OR902593) to S. solidus (accession numbers MW602517.1, MW602521.1 and AP017669.1) and there was a 98.74% similarity between 1 parasite from a ninespine stickleback (accession number OR902594) to S. pungitii (accession number MW602516.1), whereas the other 3 parasites from ninespine sticklebacks (accession numbers OR902595–OR902597) had only a 86.48% similarity with S. pungitii (accession number MW602516.1), but a 94% similarity with S. solidus (accession numbers MW602517.1, MW602521.1 and AP017669.1). Nucleotide sequences from coastrange and slimy sculpin parasites were similar (overall sequence divergence of 0.7%) whereas there was 4% sequence divergence between parasites from threespine and ninespine stickleback hosts. Notably, there was 20.5% nucleotide sequence divergence between parasites from sculpin and stickleback hosts. Amino acid similarity was 90% between the parasites from sculpin hosts and S. cotti, 85% between the parasites from sculpin hosts and S. solidus (accession numbers QXU59603.1 and QXU59651.1), and there was a 86% similarity between parasites from sculpin hosts to S. pungitii (accession number QXU59591.1).
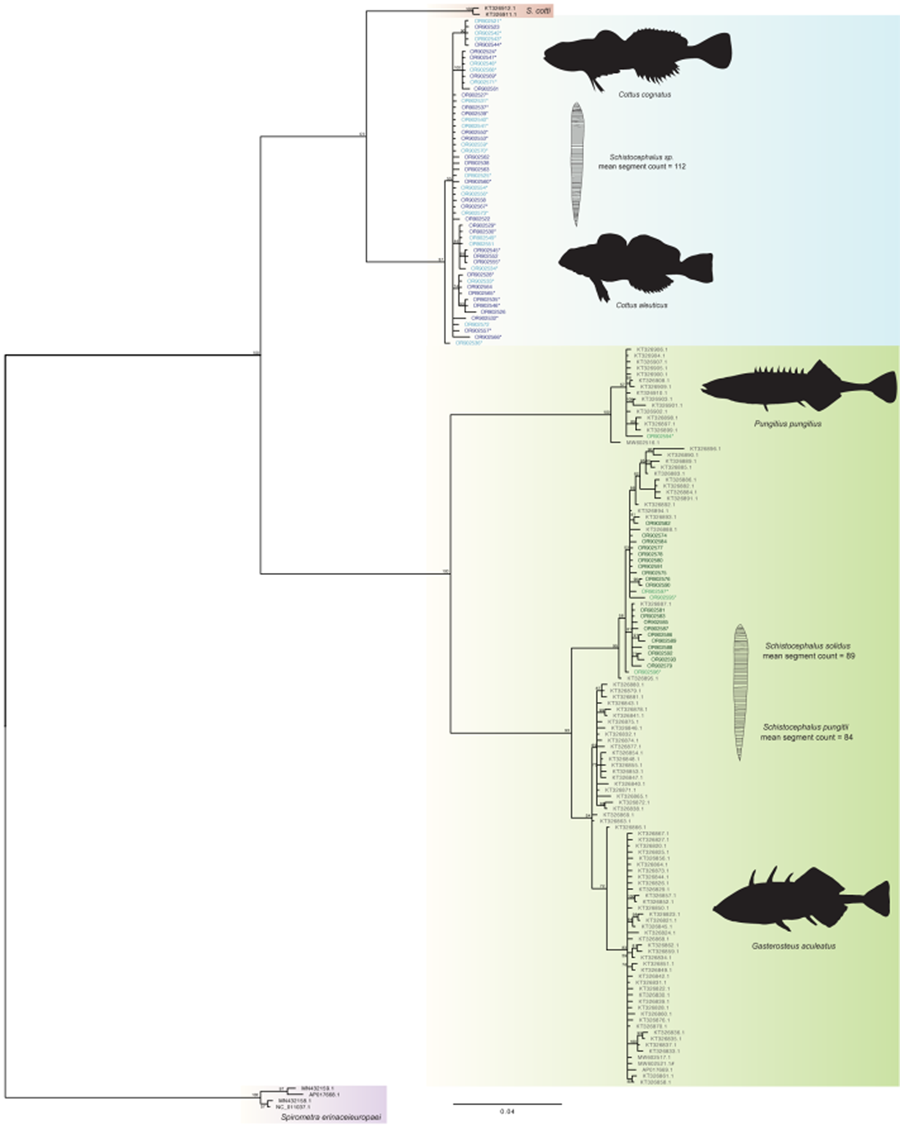
Bayesian phylogenetic analysis recovered 2 distinct clades (Fig. 2), one composed of parasites found in sculpin hosts and the other of parasites found in stickleback hosts. The 2 clades were separated by approximately 20% sequence variation without ambiguity. Neither lake nor collection year moderated the tree structure – all sculpin derived parasites clustered within the sculpin clade and likewise, all stickleback derived parasites clustered together. Support was not found for distinct clusters of parasites from threespine and ninespine stickleback hosts, respectively, nor for parasites clustering according to sculpin host species (Fig. 2).

Figure 2. Bayesian tree (scale bar: 0.02 estimated substitutions per site) of Schistocephalus parasites sequenced with partial NADH1 gene from their respective host fish species: Cottus cognatus parasites (n = 20), light blue; Cottus aleuticus parasites (n = 33), dark blue; Gasterosteus aculeatus parasites (n = 20), dark green and Pungitius pungitius parasites (n = 4), light green. * denotes corresponding segment counts were obtained from the individual; # denotes Pusa hispida botnica host. Fishes and parasites are not drawn to scale.
Discussion
Here we provide evidence of host specificity and differentiation among Schistocephalus plerocercoids infecting a complement of co-occurring host species. All 4 fish hosts are either regularly or incidentally susceptible to infection via trophic transmission within the local community. Infections conceivably could have arisen from non-specific transmission whereby all hosts were infected by the same parasite. To the contrary, our results indicate that infection is moderated by host specificity, where evolutionarily distinct Schistocephalus parasites infect different intermediate host species. Schistocephalus from sculpins differed from those in sticklebacks, supporting prior research pointing to host specificity. Chubb et al. (Reference Chubb, Seppälä, Luscher, Milinski and Valtonen2006) proposed that Schistocephalus infecting cottids are evolutionarily distinct from those in threespine sticklebacks based on significant differences in mean segment number and PCR amplification trials suggestive of nucleotide sequence divergence. We detected similar meristic differences, and our genetic and phylogenetic analyses revealed that Schistocephalus plerocercoids from cottids are highly differentiated from those in sticklebacks, bolstering the argument for recognizing Schistocephalus infecting cottids as 1 or more distinct evolutionary lineages (i.e. species). We did not recover clear evidence of finer-scale evolutionary divergence, but our findings are nonetheless broadly consistent with phylogenetic evidence that Schistocephalus diversification corresponds to host species specificity (Nishimura et al., Reference Nishimura, Heins, Andersen, Barber and Cresko2011). A phylogeny based on mtDNA sequence variation recovered distinct clades of Schistocephalus infecting threespine stickleback and ninespine stickleback, supporting the hypothesis (Dubinina, Reference Dubinina1959) that S. solidus and S. pungitii represent 2 distinct evolutionary lineages warranting species recognition. Nishimura et al. (Reference Nishimura, Heins, Andersen, Barber and Cresko2011) also found differences despite the potential for substantial gene flow among parasites in areas supporting populations of both sticklebacks, suggesting that S. solidus and S. pungitii are good biological species. Mounting additional efforts to build on our study would likely shed further light on the specificity of Schistocephalus parasites within and among intermediate fish hosts.
The morphological phenotypes of plerocercoids from the 2 stickleback species were distinct from the plerocercoids infecting the 2 sculpin species. There were significant differences in mean segment count for all comparisons within and between lakes, except for the low sample size comparison between slimy sculpin (n = 12) and threespine stickleback (n = 6) in Aleknagik Lake. Though compelling, a difference in segment counts is neither indisputable evidence of evolutionary differentiation nor can it serve as a definitive basis for taxonomic identification. Prior research has questioned the importance and use of segment counts as a diagnostic attribute. Both Clarke (Reference Clarke1954) and Dubinina (Reference Dubinina1980) concluded that segment number of fully segmented young plerocercoids exhibits little increase with further growth, and Dubinina (Reference Dubinina1980) suggested that segment number is a genetically determined trait. Chubb et al. (Reference Chubb, Valtonen, McGeorge and Helle1995), however, concluded that segment number is phenotypically plastic and related to plerocercoid size. Chubb et al. (Reference Chubb, Seppälä, Luscher, Milinski and Valtonen2006) later proposed that plerocercoid and adult segment number could be used to identify Schistocephalus species and included the trait in their taxonomic key to plerocercoids of Schistocephalus species. Further study of this trait is warranted; experimental research (e.g. a common garden experiment) to evaluate heritability and plasticity could be especially informative.
Phylogenetic analysis recovered 2 well-supported monophyletic clades, with approximately 20% nucleotide sequence divergence separating Schistocephalus infecting sticklebacks from those in sculpin hosts. Membership in the clades did not vary according to sampling location or year. The estimated percentage of divergence is widely associated with species- or higher levels of taxonomic differentiation. For example, there is only 1.24% genome-wide sequence divergence between humans and chimpanzees (Ebersberger et al., Reference Ebersberger, Metzler, Schwarz and Pääbo2002), and ~2% mtDNA sequence divergence is widely used for affirming or recognizing species of freshwater fish (Blum et al., Reference Blum, Neely, Harris and Mayden2008). We detected no ambiguous sequences between stickleback- and sculpin-derived parasite clades (no detection of any sculpin parasites in sticklebacks nor any stickleback parasites in sculpins), indicating that differentiation is not recent and that hybridization has likely not occurred between members of these 2 clades. Notably, the observed sequence variation translated to 18–20 amino acid differences between our sequenced sculpin host parasites and GenBank-derived stickleback host parasites (both 3-spine and 9-spine hosts), which offers further support for recognizing the sculpin and stickleback parasite groups as distinct evolutionary lineages. In comparison, Nishimura et al. (Reference Nishimura, Heins, Andersen, Barber and Cresko2011) proposed recognizing 2 different parasite species in threespine and ninespine sticklebacks (respectively) based on lower levels of sequence divergence. Although our phylogenetic analysis demonstrates reciprocal monophyly between parasites from P. pungitius and G. aculeatus from GenBank sequences, we did not detect a clear pattern of divergence among our parasites of the 2 stickleback species. All of our G. aculeatus sequences grouped within the S. solidus clade; however, for P. pungitius, only one of our sequences, out of four, grouped within the S. pungitii clade. This may be an artefact of analysing a relatively short region of the NADH1 gene that provided less information on sequence variation than the region examined by Nishimura et al. (Reference Nishimura, Heins, Andersen, Barber and Cresko2011). Empirical investigations to date support the conclusion that S. solidus and S. pungitii are only able to infect their respective, specific host species of stickleback (Nishimura et al., Reference Nishimura, Heins, Andersen, Barber and Cresko2011; Henrich et al., Reference Henrich, Benesh and Kalbe2013). Nonetheless, the ability to hybridize the 2 species of Schistocephalus in vivo suggests that hybridization in nature within a single host may be possible (Henrich et al., Reference Henrich, Benesh and Kalbe2013). We also did not detect a clear distinction between slimy and coastrange sculpin parasites, but we cannot exclude the possibility that the parasites comprise distinct evolutionary lineages among sculpin host species.
Further investigation focusing on these questions and on diversity among Schistocephalus parasites is warranted, particularly among parasites from sculpin hosts. Attention should also be given to Schistocephalus nemachili and Schistocephalus thomasi, which are considered valid species (Global Cestode Database), although not well studied. Our efforts were constrained in part by the utility of primers for PCR amplification and conventional Sanger sequencing. Published primer sets that work well for stickleback parasites do not perform as well for sculpin parasites. Chubb et al. (Reference Chubb, Seppälä, Luscher, Milinski and Valtonen2006) encountered similar challenges with microsatellite primers designed for Schistocephalus from threespine stickleback that did not amplify for parasites infecting bullhead, C. gobio. Accordingly, further investments should be made to develop primers and molecular markers for parasites derived from different host species. This would allow for broader sequencing of the full NADH1 gene with (putatively) lineage-specific primers. Next-generation sequencing (e.g. ddRAD single-nucleotide polymorphism analysis) could also provide greater resolution to clarify species- or population-level differences, as well as finer-scale patterns of host specificity, host–parasite evolution and trophic transmission in Schistocephalus.
Further investigation could lead to Schistocephalus being recognized and adopted as a system for studying speciation in parasites. Parasites in the diphyllobothriidean cestode genus Ligula have been the subject of more and more comprehensive investigations of evolutionary differentiation among parasites. Research thus far has revealed evidence of diversification corresponding to fish hosts and geography. Nazarizadeh et al. (Reference Nazarizadeh, Nováková, Loot, Gabagambi, Fatemizadeh, Osano, Presswell, Poulin, Vitál, Scholz, Halajian, Trucchi, Kočová and Štefka2023), for example, found strong support for 10 or more evolutionary lineages reflecting taxonomic distinctions (i.e. genera and orders) of fish hosts, including groups that differ in global extent. Differences in geographic distributions offer opportunities to study vicariant and ecological speciation among parasites (Nazarizadeh et al., Reference Nazarizadeh, Nováková, Loot, Gabagambi, Fatemizadeh, Osano, Presswell, Poulin, Vitál, Scholz, Halajian, Trucchi, Kočová and Štefka2023). As shown in previous studies (Sprehn et al., Reference Sprehn, Blum, Quinn and Heins2015; Strobel et al., Reference Strobel, Alda, Sprehn, Blum and Heins2016), S. solidus does vary genetically across different geographic regions and could explain the phylogenetic patterns within our S. solidus clade (Fig. 2). Unfortunately, geographic data are not available for the sequences obtained through GenBank that start with KT. Additional geo-referenced sampling and sequencing could help clarify these patterns, the potential drivers of genetic variation and potentially cryptic divergence. Discovering cryptic species is important to gaining greater insight into community structure and function, as well as processes of evolutionary biology and biogeography (Pérez-Ponce de León and Nadler, Reference Pérez-Ponce de León and Nadler2010; Nadler and Pérez-Ponce de León, Reference Nadler and Pérez-Ponce de León2011). Revealing crypsis through modern molecular methods is especially important for parasites that are morphologically simple with few diagnostic characteristics (Hanelt et al., Reference Hanelt, Schmidt-Rhaesa and Bolek2015), and it is even more so for morphologically simple parasites with unreliable morphological traits such as Schistocephalus. Our findings illustrate that research on Schistocephalus parasites is a potentially fertile area of enquiry using state-of-the-art molecular tools to manifest findings that complement those from ongoing research on Ligula.
In addition to the opportunities for further research on the parasites themselves, our study highlights the need for more information on the possible mode of infection of sculpins by Schistocephalus parasites. Sampling of coastrange and slimy sculpin from Iliamna Lake has not revealed any zooplankton in the diets ( P. B. Roger, unpublished MSc thesis, University of Washington, 1971; B. S. Harmon, unpublished data, 2012). A literature review of coastrange and slimy sculpin food studies from other North American lakes either did not uncover zooplankton in the diet or found it to be a very minor component. Only 1 study mentioned cyclopoid copepods (Bunnell et al., Reference Bunnell, Davis, Chriscinske, Keeler and Mychek-Londer2015). Consumption of cyclopoid copepods, the intermediate host of Schistocephalus, appears to be very limited among fish 20–100 mm standard length, the size range primarily sampled in the aforementioned studies. Other freshwater sculpin species in lakes elsewhere substantially consume cyclopoid copepods but apparently only seasonally as young-of-the-year (YOY) fish <20 mm standard length (Broadway and Moyle, Reference Broadway and Moyle1978; D. Neverman, Unpublished MS Thesis, Utah State University, 1989). Similarly, threespine stickleback become infected seasonally soon after hatching as YOY (Heins et al., Reference Heins, Eidam and Baker2016; Wohlleben et al., Reference Wohlleben, Steinel, Meyer, Baker and Foster2022). We hypothesize that coastrange and slimy sculpins also become infected seasonally soon after birth as YOY fish. Further research on the trophic ecology of sculpins, especially their consumption of zooplankton and means of infection, remains a critical area of investigation. Systematic investigations of the trophic ecology, linked to infection rates, for both sculpin species in a range of habitats would be fruitful. They occur in streams and lakes, for example, but the extent of movement between these habitats is unclear. In addition, better information on the comparative ecology (diet and habitat use patterns) of the 2 stickleback species, and the key avian predators for all these species would be informative.
In conclusion, an integrative systematic approach combining ecological, morphological and genetic data supports the hypothesis that parasites infecting coastrange and slimy sculpins in Aleknagik and Iliamna lakes of Alaska are biologically distinct, apart from the 2 known species of parasites infecting ninespine and threespine sticklebacks. Our goal was to test for these differences and to summarize what is known about the evolutionary diversification of cestodes in the genus Schistocephalus. These parasites offer a challenging and potentially enlightening investigation into adaptive radiation. For example, we do not know whether the parasites in coastrange and slimy sculpins we studied represent 2 separate species, nor whether any of those parasites differ from S. cotti. The species-level host specificity thus far observed for parasites infecting sticklebacks suggests that there may be 3 biological species infecting the sculpins known to be parasitized by Schistocephalus. The results of this investigation should inform future research and provide a foundation for detailed systematic studies of diversity and dynamics of the evolutionary pattern presented by the genus Schistocephalus.
Data availability statement
Sequence data are available in GenBank (accession numbers OR902521–OR902597) (upon publication).
Acknowledgements
Many individuals participated in the field over the years but we especially thank Jackie Carter for help at Aleknagik Lake and Curry Cunningham and Jason Ching at Iliamna Lake. We thank Haley Kodak (University of Tennessee) and Megan Sekiya (Tulane University) for molecular data collection, as well as Hannah Evans (University of Tennessee) for molecular and meristic data collection.
Author contributions
The initial observation of the cestodes in sculpins was made by B. S. H. The study was conceived and designed by M. J. B., K. N. M., D. C. H. and T. P. Q. Field sampling was conducted by M. C. A., B. S. H. and T. P. Q. Segment counts were performed by M. C. A. and B. S. H. Genetic analyses were conducted by M. J. B. and K. N. M. Statistical analyses were performed by M. C. A. and K. N. M. A draft and revisions of the manuscript were completed with contributions from all authors and coordinated by D. C. H.
Financial support
We did not receive support directly for this specific study. Instead, the study was supported indirectly by funding to the authors while this investigation was completed, including from Tulane University, the Newcomb College Institute of Tulane University and the University of Tennessee. The University of Washington's field programme in Alaska, from which this study originated, has been supported by the Pacific salmon seafood processing industry, the Gordon and Betty Moore Foundation, Alaska Department of Fish and Game, National Science Foundation, the University of Washington and other sources over the decades of the programme's existence including the sampling included here. M. C. A. was supported by the WHOI President's Innovation Fund.
Competing interests
None.
Ethical standards
Field collection of samples was approved by the University of Washington's IACUC (Protocol no. 3142-01, renewed annually) and conducted with permits from the Alaska Department of Fish and Game (e.g. for 2012: SF2-12-108d, SF2012-110d and SF2012-114d, and comparable annual renewals).