This paper contains supplementary material that can be found online at http://journals.cambridge.org
Introduction
Protecting large carnivores in the face of increasing human populations, agricultural expansion and over-use of natural resources is one of the biggest conservation challenges. Communities in the vicinity of wildlife parks and sanctuaries are reliant upon livestock for their livelihoods, resulting in increasing numbers of livestock even within protected areas (Mishra et al., Reference Mishra, Van Wieren, Ketner, Heitkonig and Prins2004). The resulting distorted relative proportions of wild and domestic prey may trigger attacks by carnivores on domestic stock (Mishra et al., 2003). The economic cost of depredation on livestock results in a negative attitude towards carnivores (Treves et al., Reference Treves, Wallace and White2009) and further aggravates human–carnivore conflicts (Graham et al., Reference Graham, Beckerman and Thirgood2005). Large carnivores also attack humans occasionally (Conover et al., Reference Conover, Pitt, Kessler, DuBow and Sanborn1995; Packer et al., Reference Packer, Ikanda, Kissui and Kushnir2005; Lodhi, Reference Lodhi2007; White & Gehrt, Reference White and Gehrt2009), resulting in injury and sometimes loss of life. The nature and extent of such conflicts seem to be more severe in South Asia (Mishra, Reference Mishra1997; Mishra et al., Reference Mishra, Allen, McCarthy, Madhusudan, Bayarjargal and Prins2003; Ogra, Reference Ogra2008) as a result of limited availability and sharing of resources.
A typical example of human–carnivore conflict in Pakistan involves the common leopard Panthera pardus. Although highly adaptable and widespread in Africa and Asia (Nowell & Jackson, Reference Nowell and Jackson1996), the leopard is threatened in Pakistan (Sheikh & Molur, Reference Sheikh and Molur2004). It has flexible habitat preferences, occupying dense forest, rock and scrub, grasslands and mountain cliffs where sufficient prey is available (Prater, Reference Prater1993; Nowell & Jackson, Reference Nowell and Jackson1996; Al-Johany, Reference Al-Johany2007). In Pakistan it is confined to Himalayan forest regions up to the limit of the tree line, and lower-altitude valleys in arid mountainous regions. It also inhabits broken, hilly terrain with Acacia scrub forest cover throughout Waziristan, Baluchistan and Sindh Kohistan (Roberts, Reference Roberts1997). The forested areas of the Himalaya, particularly the Galliat in the Khyber Pakhtunkhwa province and Neelam Valley in the state of Azad Jammu and Kashmir, are probably the main strongholds of the leopard in Pakistan. Elsewhere, the common leopard is rare, having been hunted almost to extinction by trophy hunters and in retaliatory killings by local people (Roberts, Reference Roberts1997).
Since 2000 there have been increasing reports of leopard–human conflict in the forested areas of the Himalaya, with casualties on both sides. The Khyber Pakhtunkhwa Wildlife Department reported 142 livestock losses attributed to common leopards in the Ayubia National Park (Lodhi, Reference Lodhi2007), and 363 livestock losses were reported from the Machiara National Park, Azad Jammu and Kashmir (Dar et al., Reference Dar, Minhas, Zaman and Linkie2009) during a 3-year period, indicating that the common leopard is the major livestock predator (90%) in the Park. Kabir & Waseem (Reference Kabir and Waseem2010) documented 72 livestock losses from the Pir Lasora National Park, Azad Jammu and Kashmir, comprising goats Capra hircus (46%), domestic dogs Canis lupus familiaris (32%), sheep Ovis aries (17%) and cows Bos taurus (4%). In addition to depredation on domestic livestock there has been an increase in the number of reports of fatal attacks on humans by leopards.
The Khyber Pakhtunkhwa Wildlife Department has organized a number of consultative workshops to help formulate an effective strategy for conservation of the common leopard in Pakistan and address the various challenges involved. There is a dearth of information about the population and ecology of leopards in Pakistan, and wildlife managers and conservationists have divergent opinions. Local communities attribute the increase in human–leopard conflict to an increasing leopard population, and propose trophy hunting as a solution (Lodhi, Reference Lodhi2007). However, wildlife managers argue that the assumption of population increase is not substantiated by credible data and that reports of livestock predation are exaggerated, because the leopards have sufficient wild prey available in the form of rhesus macaques Macaca mulatta.
An accurate assessment of livestock predation is necessary to measure the scale of the human–leopard conflict and design appropriate management strategies. Here we apply a DNA-based diet analysis technique (Shehzad et al., Reference Shehzad, Riaz, Nawaz, Miquel, Poillot and Shah2012b) to a population of the common leopard in Ayubia National Park. Our findings facilitate understanding of the feeding ecology of the common leopard and its conflict with the local community and will inform efforts to mitigate this conflict.
Study area
Ayubia National Park (33 km2) was established in 1984 and is surrounded by 150 km2 of reserved forests (Fig. 1). It hosts c. 200 species of birds, 31 species of mammals, 16 species of reptiles and three species of amphibians (Farooque, Reference Farooque2007). The creation of the Park has facilitated the recovery of a number of species, including the common leopard, which was close to extinction in the Galiat region and adjacent areas by the early 1980s (Lodhi, Reference Lodhi2007).
Fig. 1 North-east Pakistan, showing the location of Ayubia National Park.
Methods
We analysed leopard diet by characterizing the prey DNA present in faecal samples after amplification of a diagnostic fragment and sequencing of polymerase chain reaction (PCR) products, using next-generation sequencing. This provides diet information without any a priori knowledge about the prey and is a cost-effective method as millions of readings can be generated from a single sequencing run. This method has several advantages over classical microscopy, which requires substantial skill and time and is prone to misidentification in the case of closely related species (Pompanon et al., Reference Pompanon, Deagle, Symondson, Brown, Jarman and Taberlet2012).
Faecal samples were collected by three experienced researchers and nine park guards. The guards were experienced in identifying leopard sign, including faeces, and they also received 1 day of training before the survey. Three teams of 3–4 persons were mobilized every day during 25 June–6 July 2008 and a total of 27 transects of 2–7 km were established in the Park and surrounding reserve forest for the collection of samples. Movement in the area is restricted to available tracks but transects were placed to maximize the spatial coverage of samples. The age of all samples collected was estimated based on their physical appearance. One hundred and eleven putative leopard faecal samples were collected.
All DNA extraction was performed in a laboratory dedicated to the extraction of degraded DNA. Total DNA was extracted from c. 15 mg of faeces, using the DNeasy Blood and Tissue Kit (QIAgen GmbH, Hilden, Germany), following the manufacturer's instructions, with a slight modification as described by Shehzad et al. (Reference Shehzad, Riaz, Nawaz, Miquel, Poillot and Shah2012b). Blank extractions were performed to monitor any contamination.
The sequence analysis and taxon assignation were done using OBITools, as described in Shehzad et al. (Reference Shehzad, McCarthy, Pompanon, Purevjav, Coissac, Riaz and Taberlet2012a, Reference Shehzad, Riaz, Nawaz, Miquel, Poillot and Shahb). Full details of the primer design and DNA sequencing can be found in Supplementary Material 1 and Supplementary Table S1.
To evaluate adequacy of sample size for characterizing leopard diet we constructed a species accumulation curve and extrapolated the species richness, using the specpool function in R (R Development Core Team, 2012). This function estimates the number of undetected species and adds them to the observed species richness (Palmer, Reference Palmer1990; Colwell & Coddington, Reference Colwell and Coddington1994).
Results
Of 111 putative leopard faecal samples collected from Ayubia National Park and surroundings, 43% were ⩾ 1 month old and the remainder were fresh samples. Sixty (54%) samples were identified as of common leopard, using the common leopard specific primer pair (PantF/PantR), and selected for further experimentation. After assembling the forward and reverse reads and filtering for primers and tags we obtained a total of 652,090 sequences, corresponding to 36,929 different sequences (DRYAD, 2014). The removal of sequences shorter than 60 bp and with a total count of < 100 reduced the dataset to 150 sequences. The next step aimed to remove sequences corresponding to PCR errors. We used the obiclean algorithm, which relies on a directed acyclic graph build for each PCR, where vertices are sequences and edges link two vertices differing only by one substitution or insertion/deletion (De Barba et al., Reference De Barba, Miquel, Boyer, Mercier, Rioux, Coissac and Taberlet2014). True sequences are assumed to be ‘head’ (i.e. the most frequent sequence among all sequences that can be linked together) or ‘singleton’ (i.e. a sequence with no variant differing by a single indel or substitution). All other sequences (n = 94) were removed from the dataset. We also removed 41 sequences with a total count among all samples of < 750. This threshold was determined to avoid having the same species identified twice in the result list (a first identification corresponding to the true sequence and a second one corresponding to an artifactual sequence). Finally, three sequences without a perfect identity corresponding to the reference database were also removed, resulting in 12 (including one leopard and 11 prey sequences) molecular operational taxonomic units (Blaxter, Reference Blaxter2004). Supplementary Table S2 presents an overview of sequence counts from next-generation sequencing initial files at different stages of the analysis. The filtered data are available at DRYAD (2014). All faecal samples identified as common leopard, using the common leopard specific primer pair (PantF/PantR), were further confirmed by the sequencing, and common leopard sequences were observed in all samples, with an overall relative frequency of 0.16. This means that the 2 μM concentration of blocking oligonucleotide used in this experiment reduced but did not completely prevent amplification of the common leopard sequence. Only common leopard DNA was amplified from three samples, with no information about prey items. Eleven prey taxa were identified in the remaining 57 samples. Three prey items were identified in one sample, two prey items in seven samples, and single prey items in 49 samples. Based on the frequency of occurrence of prey items in the 57 faecal samples, the domestic goat predominated the diet (64.9%), followed by dog (17.5%) and cow (12.3%). Domestic animals (goat, dog, cow, water buffalo Bubalus bubalis, horse Equus caballus and sheep) occurred in 54 of 57 samples, corresponding to a frequency of occurrence of 0.95, and five samples contained two items of domestic prey. Fig. 2 illustrates the relative frequencies of prey items present in the leopard's diet.
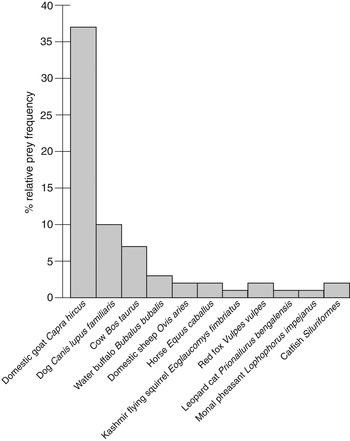
Fig. 2 Relative frequency of prey items in the diet of the common leopard Panthera pardus in Ayubia National Park, Pakistan (Fig. 1).
Main prey items (goat, dog, cow, buffalo), with cumulative frequency of 85%, were detected in the first four samples. A species accumulation curve with 100 permutations reached a species richness of 10 at sample 48, so there was an addition of only one species in the remaining 12 samples. The extrapolated species richness was 13.67 ± SD 3.48, which is close to the observed number of species (11). On this basis we believe that the sample size was adequate to characterize the diet of common leopards, although additional samples could reveal some occasionally or opportunistically eaten species.
Discussion
Analysis of faecal samples by combining universal primers for vertebrates (plus a blocking oligonucleotide specific to predator DNA) with next-generation sequencing revealed the diet of the common leopard in Pakistan. In 57 samples we identified a total of 11 prey species. Our results confirm that the common leopard is a generalist predator, consuming a wide variety of prey, including ungulates, carnivores, rodents, birds and fish.
Throughout the leopard's range there has been a shift in its prey choices towards livestock (Judas et al., Reference Judas, Paillat, Khoja and Boug2006; Spalton & Al Hikmani, Reference Spalton and Al Hikmani2006). Circa 95% of the leopard prey items in Ayubia National Park are domestic animals and there are a number of possible reasons for such a high predation rate. Firstly, the lack of natural prey in the Park provokes leopard attacks on domestic stock. Secondly, poor husbandry and penning of livestock make domestic animals more vulnerable to leopard attacks. Thirdly, seasonal factors affect the susceptibility of livestock to predation; the sampling for this study was carried out during the summer, when most of the livestock is sent to pasture for grazing and is mostly unattended. Grazing livestock is occasionally guarded by dogs, which are the second-most common leopard prey species in summer. We also found leopard cat Prionailurus bengalensis, red fox Vulpes vulpes and Kashmir flying squirrel Eoglaucomys fimbriatus in the leopard diet, which have not been reported previously as leopard prey. Among the 11 taxa predated by leopards in Ayubia only one species, the Kashmir flying squirrel, is categorized as Vulnerable in Pakistan (based on national assessment of mammals in Pakistan; Sheikh & Molur, Reference Sheikh and Molur2004). This species was present in a single sample, indicating that it is not under significant predation pressure from leopards, although it is heavily predated by the leopard cat in the same area (Shehzad et al., Reference Shehzad, Riaz, Nawaz, Miquel, Poillot and Shah2012b). As our results are based on summer diet they may not reflect the complete diet profile of the leopard in the area surveyed. Future work based on samples collected year-round could reveal more prey taxa in the leopard diet.
The rhesus macaque is common in Ayubia National Park and is generally believed to constitute a major part of the leopard diet there (Lodhi, Reference Lodhi2007). However, our findings do not support this perception as we did not find any monkey sequences in the leopard faeces despite the fact that our reference database contained several exemplars. There are conflicting reports about leopard predation on primates from different parts of its range in Asia and Africa (Kummer et al., Reference Kummer, Banaja, Aro-Khatwa, Grandour, Wittmer and Biittiker1981; Cowlishaw, Reference Cowlishaw1994; Isbell, Reference Isbell1994; Nowell & Jackson, Reference Nowell and Jackson1996; Zuberbühler & Jenny, Reference Zuberbühler and Jenny2002; Hayward et al., Reference Hayward, Henschel, O'Brien, Hofmeyr, Balme and Kerley2006). Leopards have disparate preferences for different primate species, and scientific data indicate that leopards rarely prey on rhesus macaque (Mukherjee & Mishra, Reference Mukherjee and Mishra2001). We therefore believe that reports (Lodhi, Reference Lodhi2007) of leopard predation on rhesus macaque in Ayubia are based on anecdotal information and misconception because the monkeys are abundant in the Park.
Our findings indicate that the leopard population in Ayubia is sustained by domestic animals. Actual loss of livestock may be even higher than that observed in the diet analysis because leopards are considered to be surplus killers and have been reported to kill up to 22 sheep in a single attack (Sangay & Vernes, Reference Sangay and Vernes2008). This level of predation can have adverse effects on the local economy and cause conflict between the leopard and the local community. Mitigation of this conflict is an important factor in leopard conservation in the Park.
We recommend a two-step strategy to engage the local community in conservation efforts. Immediate measures could include the introduction of incentives to foster a greater acceptance of the predator, such as support for livestock insurance (Mishra et al., Reference Mishra, Allen, McCarthy, Madhusudan, Bayarjargal and Prins2003), vaccination programmes (Nawaz, Reference Nawaz2009) and improving livestock-guarding facilities (Dar et al., Reference Dar, Minhas, Zaman and Linkie2009). Such incentives could be subsidized from the revenue generated by Park entry fees and associated tourism.
Measures to curtail predation can also help reduce conflict and can be complemented by incentive programmes. For example, improvements in penning and guarding facilities can result in significant reductions in livestock losses to predation. Increased vigilance at night and guarding livestock during grazing can reduce the risk of leopard attacks (Dar et al., Reference Dar, Minhas, Zaman and Linkie2009).
A second step should focus on the reintroduction and protection of wild prey in the Park. Depletion of natural prey as a result of poaching is the main reason for the leopard's dependence on livestock for food and is thus a major source of conflict. Reintroduction of wild prey species such as goral Naemorhedus goral and musk deer Moschus moschiferus could help resolve these issues in the longer term. The Wildlife Department's existing captive facility at Nathia Gali could be used for breeding of potential prey species before release in the wild. However, such efforts would only be successful if poaching of these species in the Park and surrounding areas could be curbed through strict law enforcement.
We hope that our results will stimulate the implementation of such a conservation strategy and ensure the survival of the leopard population in Pakistan. The results of our study were shared informally with the Wildlife Department and conservation organizations working in the area, which promoted a realization that communities’ concerns over livestock predation are real and significant. Consequently, programmes have been initiated to compensate communities for their losses.
Acknowledgements
The authors are grateful to Alice Valentini, Christian Miquel, Carole Poillot, Delphine Rioux and Ludovic Gielly for their valuable comments and suggestions throughout this study. Wasim Shehzad and Tiayyba Riaz were supported by PhD scholarships from the Higher Education Commission of the Government of Pakistan. The Khyber Pakhtunkhwa Wildlife Department, Himalayan Wildlife Foundation and Snow Leopard Trust organized sample collections in Ayubia National Park.
Biographical sketches
Wasim Shehzad's research interests cover ecological studies exploiting DNA from non-invasive sampling, and applying this approach to biodiversity surveys and the identification of elusive and threatened species. Ali Nawaz is interested in molecular and field studies for natural resource management, and has been focusing on understanding ecology, co-existence and conservation issues of the carnivore community in Pakistan. François Pompanon studies adaptation through population genomics approaches and develops molecular markers for describing biodiversity. Eric Coissac and Tiayyba Riaz are involved in developing the DNA metabarcoding approach, focusing on the bioinformatics aspects. Safdar Ali Shah is experienced in collaborative management of biodiversity and its sustainable use through community participation. Pierre Taberlet is interested in conservation genetics and develops molecular methods (including metabarcoding) for studying non-invasive samples.




