Introduction
Epigenetics has been defined as a series of reversible and inheritable alterations capable of regulating genetic expression and stability. These mechanisms support cellular growth, development and differentiation, but they do not disrupt DNA sequences of bases(Reference Mazzone, Zwergel and Artico1). The epigenetic settings may be orchestrated by several factors such as the aging process or environmental influences such as smoke, climate, nutrition, viral infections, chemical exposures, etc.(Reference Costenbader, Gay and Alarcón-Riquelme2). Three main epigenetic mechanisms involved are DNA methylation, histone modifications and noncoding RNA profiling (Fig. 1).
-
1. DNA methylation. DNA methylation is the most traditional and well-known epigenetic event. It is involved in several pathologies, considered as a basic process of cellular development and growth. This procedure involves the transference of a methyl group from S-adenosylmethionine (SAM) to 5′-carbon position of cytosine residues in cytosine-phosphate-guanosine (CpG) dinucleotides on DNA strand. The CpG islands are located near promoter regions and transcription start sites. For that reason, the methylated CpGs sites result in inaccessible heterochromatin for transcription effectors, inducing genic silencing. Contrarily, the non-methylated sequences remain accessible, and transcription goes on(Reference Aslani, Mahmoudi and Karami3,Reference Foma, Aslani and Karami4) . DNA methylation is executed by DNA methyltransferase (DNMT) enzymes. According to de novo or maintenance states of methylation, it may be conducted by DNMT3 or DNMT1, respectively.
-
2. Histone modifications. Histones are proteins that wrap DNA to form nucleosome structures. The main post-translational alterations of histones are acetylation/deacetylation, methylation/demethylation, phosphorylation and ubiquitination. These modifications at terminal amino acid modulate the accessibility to transcriptional factors and gene expression(Reference Araki and Mimura5). Some studies have postulated a relation between DNA methylation and histone modifications that reorganises chromatin and blocks gene expression(Reference Hedrich6).
-
3. Noncoding RNAs (ncRNAs). Long noncoding (lnc) (>200 nucleotides) and short ncRNAs (miRNA) (<200 nucleotides) modulate expression or repression at the transcriptional and post-transcriptional level, regulating chromatin formation, histone modifications and DNA methylation. miRNAs have been extensively studied in autoimmune and chronic inflammation, being postulated as biomarkers and therapeutic targets. In addition, miRNAs bind to complementary target mRNAs and suppress translation by degradation of this target(Reference Meda, Folci and Baccarelli7).
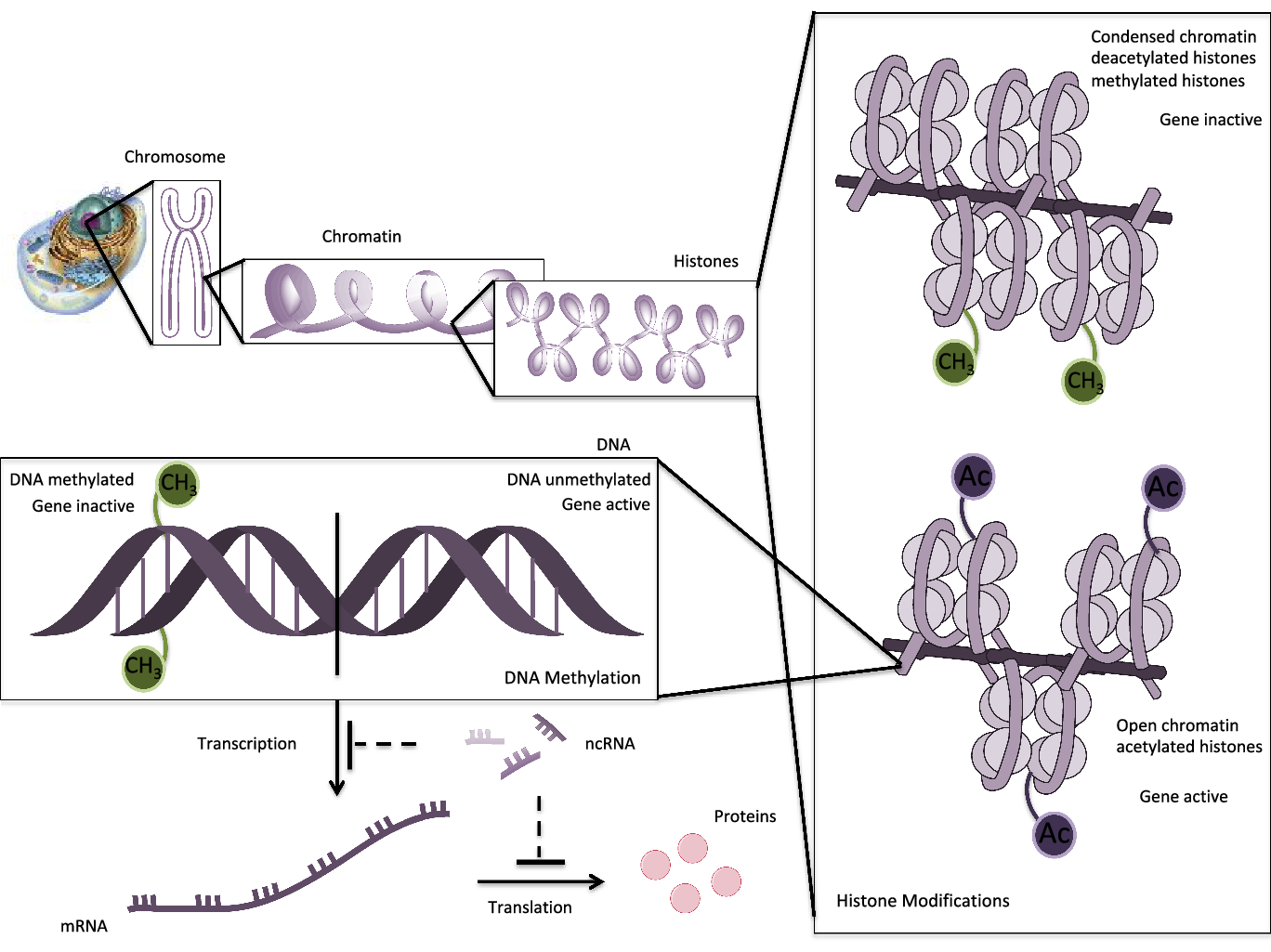
Fig. 1. Schematic epigenetic modifications: histone modifications, DNA methylation and ncRNAs. DNA is wrapped around histones to form nucleosomes. Histone acetylation opens the chromatin, enabling transcription with activation of genes. Contrarily, deacetylation or methylation of histones condenses the chromatin, making it inaccessible for transcription components. DNA methylation is operated by DNMTs that add methyl groups on CpG island in the promoter region of a gene. DNA methylation blocks the binding of transcription factors and results in suppression of gene expression. In unmethylated DNA sequences, the transcription machinery is capable to transcribe and gene is active. ncRNAs regulate gene expression or silencing at the transcriptional or post-transcriptional level.
The maintenance of proper function and balance between epigenetic processes is essential to normal development and action of immune system. However, increasing evidence has shown that epigenetic deregulated modifications, including DNA methylation, histone modification and ncRNAs, are involved in the pathogenesis of several autoimmune diseases (AD). Besides, the alterations of epigenome are implicated in the dysregulation of signalling molecules and receptors of various autoimmune/inflammatory conditions(Reference Hedrich6,Reference Jeffries and Sawalha8) . Also, it has been reported that ADs are related to genetic predisposition and environmental factors, whose effects induce epigenetic modifications(Reference Ngalamika, Zhang and Yin9).
Consequently, epigenetic events could play a relevant role in the aetiology and pathogenesis of ADs, even promising a potential future therapeutic intervention. In fact, it has been proposed as a plausible strategy in the research of new therapeutic agents that are more specific and personalised. Thus, pharmacologic and pharmaco-epigenetic studies on reversible epigenetic regulations of autoimmune pathologies to drug responses may provide novel individualised therapies(Reference Aslani, Mahmoudi and Karami3,Reference Jeffries and Sawalha8,Reference Ngalamika, Zhang and Yin9) .
ADs like systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), psoriasis or multiple sclerosis, among others, are characterised by epigenetic marks in affected cells(Reference Aslani, Mahmoudi and Karami3). Despite the fact that total extension of epigenetic modifications in these pathologies remains unclear, this characteristic could indicate a definitive approach to patients who are exposed to chronic therapy whose side effects can make day-to-day life difficult for patients(Reference Meda, Folci and Baccarelli7).
Overall, nutrition is one of the most accessible external factors implicated in ADs, so its influence on the epigenetics profile of ADs should be considered. Additionally, the knowledge of micronutrient action in epigenetic patterns of ADs could indicate a promising strategy to prevent them. Nevertheless, the studies that investigate the effects of bioactive compounds in the context of epigenetic alterations have been focused mainly on cancer. Consequently, in the present review, we intend to summarise up-to-date preclinical and clinical studies on epigenome alterations of SLE and discuss the potential roles of epigenetic regulation by dietary bioactive compounds.
Systemic lupus erythematosus (SLE)
SLE is an autoimmune multiorgan disease characterised by its clinical and pathogenic complexity that can damage different organs such as skin, kidneys, joints, lungs, coronary system and liver, among others(Reference Aparicio-Soto, Sánchez-Hidalgo and Alarcón-de-la-Lastra10,Reference Zhan, Guo and Lu11) . Difficult diagnosis, alternated and unpredictable exacerbation and remission periods during the course of disease, in addition to the high number of complications, affect the patient’s quality of life.
This AD is characterised by loss of tolerance to self-antigens and, thus, overactivation of B and T cells, increasing the production of anti-nuclear autoantibodies, immune complex deposition, lymphoproliferation and expression of inflammatory cytokines that may initiate and amplify inflammation, contributing to the clinical manifestations of SLE(Reference Honarpisheh, Köhler and von Rauchhaupt12).
SLE is considered a rare disease, according to EC Regulation on Orphan Medicinal Products. Demographically, the prevalence of the disease is approximately 20–150 cases per 100 000 individuals worldwide. Besides, approximately 90 % of the patients are fertile women between 20 and 30 years old, although it can appear at any age (Reference Vina, Utset and Hannon13,Reference Mina and Brunner14) .
Even though aetiopathogenesis of SLE is not totally defined, genome-wide association studies have revealed several roles to genetic predisposition in lupus patients. However, the discordance for SLE between monozygotic twins described by Deapen et al., in 1992, suggested that other non-genetic factors possibly regulate disease onset (Reference Deapen, Escalante and Weinrib15). Exposure to UV light, smoking, consumption of some drugs or inadequate dietary habits could negatively condition the epidemiology of SLE (Reference Aparicio-Soto, Sánchez-Hidalgo and Alarcón-de-la-Lastra10).
Recently, epigenetics has been postulated as an environmental driver that contributes to the aetiology of SLE. In 2010, Javierre and colleagues revealed for the first time that monozygotic discordance on SLE features widespread alterations in DNA methylation status of several genes. Individual analysis confirmed different profiles of methylation and expression in genes related to SLE pathology (Reference Javierre, Fernandez and Richter16). As a result, discordance between twins allowed the identification of epigenetic targets and highlights their potential contribution to SLE.
The presence of X chromosomes is positively related to high risk for SLE. Nevertheless, the implicated epigenetic features remain unclear. Recently, Syrett and colleagues have established that the sticking female predilection of this disease is caused by the defective inactivation of X chromosome. Females normally inactivate one X chromosome to balance X-linked genes with the male sex (Reference Syrett, Paneru and Sandoval-heglund17). The inactivation process is started in early development, where each female cell randomly determines to silence the paternal or maternal X chromosome, by allele-specific up-regulation of the long noncoding RNA (lncRNA) Xist from the future inactive X. Xist RNA recruits heterochromatin modifications (H3K27me3 and H2-ubiquitin) across the X chromosome, inducing a transcriptional silencing. They observed that Xist RNA disappeared from the X inactive chromosome, and X chromosome inactivation maintenance was altered in the maturation of thymocytes and T-cell subsets from SLE patients and mice. Consequently, SLE patients’ T cells exhibited up-regulated X-linked genes(Reference Syrett, Paneru and Sandoval-heglund17). Accordingly, the hypomethylation of several genes encoded to X chromosome, such as TRL7 or CD40L, which are implicated in the development of immune response, have been described in lupus disease and pathophysiology. In fact, CD40LG, a B-cell co-stimulatory molecule, is overexpressed in lupus lymphocytes and contributes to an exacerbated production of autoantibodies (Reference Aslani, Mahmoudi and Karami3,Reference Foma, Aslani and Karami4,Reference Hedrich6,Reference Lu, Wu and Tesmer18) .
Variations of epigenome on SLE
DNA methylation, histone modifications and miRNA expression have been thoroughly defined in in vitro and in vivo experimental studies, using peripheral blood mononuclear cells (PBMCs) of SLE patients or animal models, that have shown that variation of the epigenome may lead to the onset of SLE.
DNA methylation in SLE
Recent studies have confirmed that DNA hypomethylation is a dominant factor in SLE (Table 1). In consequence, some demethylating agents are known to promote drug-induced experimental lupus (Reference Zhou and Lu19).
Table 1. DNA methylation altered patterns in SLE

Lupus-like autoimmunity and severity is associated with CD70 (so TNFSF7 encoded) and C11a (so TNFSF5 encoded) overexpression in CD4+ T cells. This fact has been related to DNA hypomethylation on their respective promoters. CD70 is expressed in activated T cells and increases IgG synthesis, collaborating in B-cell co-stimulatory functions (Reference Zhao, Sun and Gao20). On the other hand, CD11a (ITGAL encoded) is an integrin implicated in co-stimulation and cellular adhesion and also related to leucocyte function-associated antigen (LFA) 1. Increased LFA1 in T cells from lupus patients involves an autoreactive phenotype of lupus (Reference Long, Yin and Wang43). Zhao et al. (2010a) established that the overexpression of CD70 and CD11a in SLE CD4+ T cells was the result of the DNMT1 recruitment at the CD70 and CD11a promoter region, orchestrated by regulatory factor X (RFX) 1. RFX1 works like an immune-suppressor and is usually decreased in SLE patients’ cells, so it seems to be involved in lupus pathogenesis (Reference Zhao, Sun and Gao20). Conclusively, regulation of RFX1 in lupus patients provides a valuable insight into methylation-dependent gene expression on CD70/CD11a axis, which promotes autoimmune response. Furthermore, Luo et al. (2010) compared CD70 expression levels and methylation status of CD70 promoter region in CD4+ T cells from patients with subacute cutaneous lupus erythematosus (SCLE) and healthy controls. The results showed a CD70 surface-overexpressed and CD70-demethylated promoter region in CD4+ T cells from SCLE patients compared with cells from healthy subjects, concluding that demethylation of regulatory elements could contribute to the increase of CD70 expression in patients with SCLE (Reference Luo, Zhao and Lu21).
Surprisingly, an interesting in vitro study showed reduced expression of RFX1 in CD4+ T cells from patients with SLE, leading to interleukin (IL) 17A overexpression through decreased DNA methylation. The results revealed that RFX1 deficiency increased the differentiation of naïve CD4+ T cells into Th17 cells. Furthermore, the same researchers demonstrated that conditional deletion of RFX1 in mice exacerbated experimental pristane-induced lupus-like syndrome and increased Th17 cell induction. These data revealed that RFX1 functions downstream of the signal transducer and activator of transcription (STAT) 3 and phosphorylated-STAT3, inhibiting their expression, highlighting a non-canonical pathway that regulated differentiation of Th17 cells (Reference Zhao, Tan and Peng25).
Additionally, previous studies have investigated inhibition of DNMT1 activity, confirming the DNA hypomethylation profile is characteristic in lupus cells. Deng and co-workers have shown that T cells from patients with active lupus had diminished DNMT mRNA levels decreasing the Ras–mitogen-activated protein kinase (Ras-MAPK) signalling. The inhibition of signalling through the Ras-MAPK pathway with a soluble inhibitor of MAPK ERK I (MEK1) decreased DNMT mRNA and their enzyme activity, similarly to DNA hypomethylation in SLE T cells. These results suggested that a decrease in Ras-MAPK pathway signalling could be involved in down-regulation of DNMT activity in association with DNA hypomethylation in patients with lupus (Reference Deng, Kaplan and Yang31). In addition, transgenic mouse with impaired ERK pathway signalling in T cells exhibited low expression of DNMT1 and up-regulation of autoreactivity-related methylation-sensitive genes (CD70, CD11a), increased anti-dsDNA antibodies levels and glomerulonephritis (Reference Sawalha, Jeffries and Webb44–Reference Gorelik, Sawalha and Patel46).
Furthermore, recent studies have confirmed that a catalytic subunit of protein phosphatase 2A (PP2Ac), overexpressed in SLE T cells, is involved in the alteration of DNMT1 activity by the impairment of T-cell phosphorylation of MEK/ERK signalling pathways. Sunahori et al. investigated DNA methylation patterns in the PP2Ac promoter region in SLE T cells, compared with normal T cells. Specifically, in T cells from patients with active SLE, the suppression of PP2Ac increased phosphorylation of signalling pathway upstream of DNMT1 expression and activity and, thus, suppressed CD70 and CD11a expression. They concluded that PP2Ac may be a key target to control the methylation of sensitive genes in SLE T through dephosphorylation of MEK/ERK signalling pathway and activation of DNMT1 (Reference Sunahori, Juang and Kyttaris30,Reference Sunahori, Nagpal and Hedrich47) .
However, although there are many clinical studies that support the role of DNA methylation in the pathogenesis of lupus, there are only a few in vivo studies of DNA methylation implication in the development of this autoimmune disorder. In 2007, Sawalha et al. studied the existence of correlation between age-dependent autoimmunity in MRL-lpr mice and the decreased expression of DNMT1 and, consequently, T-cell DNA hypomethylation. They found lower levels of DNMT1 steady state in 16-week-old MRL-lpr mice compared with 5-week-old mice. Also, they described that 16-week-old mice presented hypomethylated CpG pairs compared with 5-week-old MRL-lpr mice (Reference Sawalha and Jeffries48). As a consequence, they concluded that reduced expression of DNMT1 and the corresponding T-cell DNA hypomethylation were correlated directly with the development of age-dependent autoimmunity in MRL/lpr mice.
Additionally, Hedrich et al. investigated CpG-DNA methylation patterns in conserved noncoding sequence regions and proximal promoter region of human IL17F gene in resting and activated naïve CD4+ T lymphocytes from healthy controls or total T lymphocytes from SLE patients. They observed that SLE T cells displayed lower degrees of CpG-DNA methylation as compared with control T lymphocytes in response to T cells. This fact suggested that IL17F production in human T lymphocytes could be regulated by CpG-DNA methylation (Reference Hedrich, Rauen and Kis-Toth23). In this sense, these researchers also studied epigenetic regulators to diametric expression of IL2 and IL17A, whose tight balance is essential for immune homeostasis. They demonstrated that cAMP response element modulator (CREM) α contributed to epigenetic remodelling of the IL2 and IL17A genes during T-cell differentiation. Furthermore, the epigenetic modulation of IL2 through DNMT3a recruitment resulted in trans-activation and demethylation of IL17A promoter. This imbalance between IL2 and IL17A expression levels is a hallmark characteristic of autoimmune disorders, such as SLE (Reference Hedrich, Crispin and Rauen24).
Other targets analysed to show the potential role of DNA methylation in the regulation of gene expression in SLE were IL4 and IL6 transcripts. Mi et al. (2008) studied in T cells isolated from SLE patients and healthy controls changes of DNA methylation in IL4 and IL6 promoters. They demonstrated that levels of IL4 and IL6 mRNA transcripts were significantly higher in SLE T cells, as compared with controls. Therefore, hypomethylation of both promoters occurred in T cells from SLE patients and was associated with the severity of SLE (Reference Mi and Zeng22).
IFI44L is considered a highly sensitive and specific diagnostic marker for SLE. Zhao et al. (2016a) found that methylation level of IFI44L promoter could distinguish patients with SLE from healthy people and other ADs; thus, it could be considered a sensitive and specific diagnostic marker for this pathology. Furthermore, the DNA methylation level within the IFI44L promoter was significantly lower in patients with SLE with renal involvement than in SLE patients without renal abnormalities (Reference Zhao, Zhou and Zhu26). In this line, stimulatory and killer cell inhibitory immunoglobulin-like receptor (KIR3) was overexpressed in T cells unmethylated from SLE patients (CD28+ and CD28−), proportionally to degree of disease, through promotion of killing of macrophages and increase in interferon (IFN) γ level, suggesting that antibodies to inhibitory KIR may be a treatment for this disease (Reference Basu, Liu and Wu49).
Overall, the wide study of overexpression of different genes such as CD11a and KIR from CD4+ and CD28+ T cells might explain genetic risk and disease activity in lupus. In fact, CD4+CD28+KIR+CD11ahi T cells are demethylated and characterised by pro-inflammatory epigenetic and transcriptional profiles in SLE. Thus, it was suggested that removing these cells or blocking their pro-inflammatory outlines could be considered a novel approach to treatment of lupus (Reference Gensterblum, Renauer and Coit28).
On the other hand, Zhao and colleagues (2016) detected increased 5-hydroxymethylxytosine (5-hmC) levels, a novel discovered modified form of cytosine, in genomic DNA in CD4+ T cells from SLE patients, compared with healthy controls. This result was accompanied by up-regulated expression of ten-eleven translocation (TET)-2 and TET-3, which can enzymatically convert 5-methylcytosine (5-mc) into 5-hmC. Also, these researchers found differences between DNA hydroxymethylation patterns from SLE patients and healthy controls, such as Wnt, MAPKs or mTOR signalling pathways and selected immune-related genes (SOCS1, NRF2F6 and IL15RA). Finally, they concluded that hydroxymethylation is implicated in aberrant regulation of genes in pathogenesis of SLE; thus, 5-hmC could provide a new tool for treating this disorder (Reference Zhao, Wang and Liao27).
Conclusively, these recent advances have shown that altered DNA methylation patterns have a plausible potential as biomarkers for diagnosis and outcome assessment, as well as objects for target and individualised interventions in SLE (Reference Hedrich, Mabert and Rauen50).
Histone modifications in SLE
Histones have octamer forms with two of each histone core (H2A, H2B, H3 and H4), which wrap DNA around them. This conformation enables the accessibility to transcriptional factors and gene expression to be controlled (Reference Hedrich6,Reference Hedrich and Tsokos51,Reference Hedrich, Crispin and Tsokos52) . Until now, the main histone modifications studied in SLE are acetylation and methylation. Both processes are mediated by different enzymes with opposite and reversible functions (Reference Wardowska, Komorniczak and Bullo-Piontecka53). In the acetylation process, an acetyl group is transferred to lysine residue on N-terminal tail by histone acetyltransferase (HAT) enzymes (Reference Hedrich6). This modification promotes an accessible chromatin structure, increasing the accessibility of DNA to transcription factors. For this reason, H3 and H4 hyperacetylation is commonly associated with gene activation. Contrarily, histone deacetylases (HDACs) catalyse the reverse process, removing acetyl groups and contributing to gene silencing through the condensation of nucleosomes and compaction of chromatin structure (Reference Foma, Aslani and Karami4,Reference Ropero and Esteller54,Reference Kim, To and Seki55) . Related to histone methylation, this can affect lysine and arginine residues (Reference Cheung and Lau56). Histone methyltransferases can add up to three methyl groups. However, depending on methylation degree and residues involved, gene regulation could be up-regulated or repressed (Reference Foma, Aslani and Karami4,Reference Zhan, Guo and Lu11,Reference Wardowska, Komorniczak and Bullo-Piontecka53) . For example, it has been described that hypermethylation of histone 4 lysine 4 trimethylation (H4K4me3) is associated with gene transcription, while H3K4me3 presents a repressive effect (Reference Long, Yin and Wang43).
Interesting studies have established that histone modifications contribute to SLE pathogenesis (Table 2). In fact, some of them have identified aberrant global methylation or acetylation of H3 and H4 in SLE patients compared with healthy controls. Nevertheless, the involvement of this epigenetic process in SLE remains unclear.
Table 2. Histone alterations in SLE

Monocytes are implicated in atherosclerosis and renal disease, contributing to mortality in SLE patients. Moreover, monocyte dysfunction induces production and response to IFN. In monocytes from lupus patients, global H4 acetylation of several genes has been reported to be significantly altered (Reference Leung, Shi and Maurer59,Reference Zhang, Song and Maurer62,Reference Leung, Maurer and Song63) . However, more than 50 % of H4-hyperacetylated genes are related to interferon regulatory factor (IRF) 1. IRF1 is a transcription factor in antiviral and immune response with anti-tumour effects. IRF1 can be induced by IFNs and tumour necrosis factor (TNF)-α, suggesting that IFN contributes to SLE pathogenesis (Reference Araki and Mimura5,Reference Kroger, Koster and Schroeder85) . Sullivan et al. (2007) revealed up-regulated ratios of H4ac in SLE monocytes at the TNF-α locus compared with healthy controls. They concluded an increased transcription of TNF- α, playing a key role in the inflammatory response that is seen in lupus patients (Reference Sullivan, Suriano and Dietzmann57).
The raised possibility that decreased histone acetylation might contribute to lupus pathogenesis by promoting silencing of some genes has been amply studied. García et al. (2005) described for the first time a global site-specific hypermethylation (except H3K4 methylation) and hypoacetylation in histone H3 and H4 MRL-lpr/lpr mice in comparison with control MRL/MPJ mice (Reference Garcia, Busby and Shabanowitz83). In agreement, Hu et al. showed that SLE CD4+ T cells had decreased overall acetylation of both H3 and H4, and they correlated the negative relation between the degree of H3 hypoacetylation with SLE disease activity (Reference Hu, Qiu and Luo75).
In early stages of lupus nephritis (LN), Wardowska and colleagues observed a significant decrease in H3K4me3 and H3K29me3 marks in dendritic cells that could reveal renal involvement in SLE, serving as distinctive biomarkers in the diagnosis of lupus nephritis and monitoring of renal outcome in SLE patients (Reference Wardowska, Komorniczak and Bullo-Piontecka53). In this line, H3K4me3 has been recognised as a potential biomarker associated to SLE pathogenesis, and it could be a promising target for epigenetic-based lupus therapies.
CREM α alters epigenetic conformation of cytokine gene by way of histone acetylation in active SLE T cells. HDAC is recruited to CRE sites in the IL2 promoter, and IL-2 expression is repressed (Reference Kyttaris, Wang and Juang86–Reference Zhang, Ding and Zhang88). Moreover, hyperacetylation of histone 3 lysine 18 (H3K18) and hypomethylated histone 3 lysine 27 (H3K27) at IL17A-IL17F promoters induce low levels of IL2 and IL17F, while IL17A is overexpressed (Reference Hedrich, Rauen and Kis-Toth23,Reference Rauen, Hedrich and Juang70,Reference Zhang, Ding and Zhang89) . Hedrich et al. (2017) agreed with these data and reported an increase in H3K27me3 and poor H3K18ac levels at IL2 promoter that silenced IL2 secretion (Reference Hedrich, Mabert and Rauen50).
Liu and colleagues studied in vitro TLR-2-stimulated CD4+ T cells from SLE patients, and they observed an increment of trimethylation into histone 3 lysine 4 (H3K4me3), and H4 acetylation together with trimethylated histone 3 lysine 9 (H3K9me3) decreased in the IL17A-IL17F promoter region (Reference Liu, Liao and Zhao73). These data could confirm that histone modifications in SLE through TLR-2 stimulation promoted IL17A and IL17F expression, developing immune reactivity. Likewise, Apostolidis et al. (2013) reported that PP2Ac, a serine/threonine phosphatase highly expressed in SLE cells, enhanced IL17 gene expression thought H3ac in murine T cells, predisposing glomerulonephritis affection (Reference Apostolidis, Rauen and Hedrich84).
IL10 is the second cytokine with enhanced expression correlated with disease activity and antibody production in lupus patient’s serum and tissues. Hedrich and colleagues investigated STAT3- and STAT5-mediated trans-activation and epigenetics remodelling of IL10 gene related to HAT p300. They concluded that the enhanced activation of STAT3 through transcriptional cofactor p300 produces a competitive replacement of STAT5 in regulatory regions and, consequently, the promotion of IL10 expression (Reference Hedrich, Rauen and Apostolidis74).
TNF-α-induced protein 3 (TNFAIP3) is a key SLE susceptibility gene implicated in the modulation of inflammatory responses through nuclear factor-κB (NF-κB) pathway. Zhao et al. studied TNFAIP3 expression in CD4+ T cells and the molecular mechanism underlying TNFAIP3 regulation in the pathogenesis of SLE, suggesting that the down-regulation of TNFAIP3 in CD4+ T cells of SLE was probably regulated by demethylation of H3K4, which led to a reduced quantity of H3K4me3 in the promoter of the TNFAIP3 gene. The dysregulation of TNFAIP3 in CD4+ T cells could play a role in the pathogenesis of SLE by over-production of inflammatory cytokines IFN-γ and IL17. Thus, they conclude that TNFAIP3 may provide a promising target for the treatment of SLE in clinical practice (Reference Zhao, Wang and Luo77).
As aforementioned, the expression and activity of the transcription factor RFX1 were decreased in SLE CD4+ T cells. Zhao and colleagues demonstrated that RFX1 affected DNA methylation and histone acetylation in CD4+ T cells by recruiting the co-repressors DNMT1 and HDAC1 to the CD11a and CD70 promoters, and thereby suppressed their expression. Reducing RFX1 in CD4+ T cells was enough to induce lupus-like T- and B-cell hyperactivity, whereas overexpressing RFX1 blocked T-cell reactivity. These findings confirm a crucial role for RFX1 in regulating the epigenetic status of T cells, and reveal that autoimmune responses in SLE are responsible in part to RFX1 down-regulation. Moreover, these authors showed that reduced expression of the transcription factor RFX1 in CD4+ T cells from patients with SLE led to IL17A overexpression through increased histone H3 acetylation and decreased DNA methylation and trimethylated H3 lysine 9 (H3K9me3) (Reference Zhao, Sun and Gao20,Reference Zhao, Tan and Peng25) .
Similarly, H3 acetylation and di-methylated H3 lysine 4 (H3K4me2) levels were significantly augmented in patients with lupus, and both factors were associated with the severity of the disease. Thus, these results showed that aberrant histone modifications within the CD70 promoter could participate in the development of lupus by increasing CD70 expression in CD4+ T cells (Reference Zhou, Qiu and Luo65,Reference Wu, Chang and Lu66) .
Hematopoietic progenitor kinase 1 (HPK1) is a negative regulator of T-cell-mediated immune responses that could play a role in lupus pathogenesis. Using chromatin immune-precipitation (ChIP) microarray data, Zhang and colleagues (2011) found significantly increased H3K27me3 enrichment at the HPK1 promoter of SLE CD4+ T cells in comparison with controls. Consistent with these findings, overexpressing HPK1 in SLE CD4+ T cells induced an important reduction in T-cell reactivity. These data could demonstrate that HPK1 may serve as a novel target for effective SLE therapy (Reference Zhang, Liao and Zhao64).
ncRNAs in SLE
ncRNAs regulate gene expression or silence at the transcriptional and post-transcriptional level. miRNAs regulate about 60 % of mRNA, and they are implicated in several ADs such as SLE or AR, among others, whereas lncRNAs act on regulated complex of development and differentiation from different immune cells through expression of active protein (Reference Mazzone, Zwergel and Artico1). Currently, they are considered excellent biomarkers and therapeutic targets in ADs (Reference Shen, Liang and Tang90,Reference Yan, Yim and Lu91) .
Several preclinical and clinical studies have investigated the role of miRNAs in lupus pathogenesis; however, thus far there has not been much study of lncRNAs. Thus, we have revised the main ncRNA epigenetic modifications described in SLE (Table 3).
Table 3. Deregulated non-coding RNAs epigenetic modifications in SLE

Among the main miRNAs postulated as biotargets and related with SLE pathogenesis were miR125a, miR142-3p/5p, miR155, miR21 and miR148, among others. Epigenetic evidence suggests that miR125a expression is decreased in monocytes and PBMCs of SLE patients. In activated T cells, miR125a targets Kruppel-like factor (KLF) 13 and then contributes to regulating the expression of ‘regulated upon activation, normal T cell expressed and secreted’ (RANTES), also known as CCL5, a known chemokine associated to inflammatory response (Reference Zhao, Tang and Qu67). Pan et al. (2015) demonstrated that miR-125a was down-regulated in peripheral CD4+ T cells of SLE patients, acting as a key regulator that controls T-cell differentiation suppressing STAT3, IL13 and IFN-γ, several gene factors in CD4+ T cells from miR125a-deficient mice (Reference Pan, Zhu and Dai92). In addition, Smith and colleagues described that IL16 was a direct target for miR125a and observed reduced pulmonary miR125a and enhanced IL16 expression, suggesting the IL16/miR125a axis as a novel therapeutic target for management of acute lung injury in SLE (Reference Smith, Wu and Seo93).
miR146a and miR155 are two of the ncRNAs most involved in ADs through NF-κB and IFN-dependent conditions. miR146a is a negative regulator of innate immunity that targets interferon regulatory factor (IRF) 5, STAT1, tumour necrosis factor receptor associated factor (TRAF) 6 and IL1 receptor associated kinase (IRAK) 1, which are crucial elements of type I IFN signalling cascade and signal transducers in NF-kB, respectively. In PBMC of SLE patients, miR146a directly repressed the transactivation downstream of type I IFN. Also, inclusion of miR146a into the patient’s PBMCs reduced the synchronised activation of the type I IFN pathway (Reference Smith, Wu and Seo93). In this context, Dai et al. (2008) observed a clear connection between miR146a expression and oestrogen production, related to the higher female prevalence of SLE (Reference Shen, Liang and Tang90,Reference Dai, Phillips and Zhang98) .
On the other hand, miR155 regulates B-cell activation and survival, essential parameters for SLE pathogenesis. Aboelenein et al. (2017) defined PU.1, a regulator of B cells that enhances TNF-α production, as a target for miR155 in PBMC and B cells from paediatric SLE patients (Reference Aboelenein, Hamza and Marzouk95). Furthermore, in vitro and in vivo studies carried out by Xin and colleagues revealed another depleted target gene of miR155, the sphingosine-1-phosphate receptor (S1PR) 1. They observed that miR155 deficiency diminished SLE autoimmune inflammation by targeting S1PR1 gene in Faslpr/lpr mice.
Complementarily, the serum levels of IL4 and IL17A, released by Th2 and Th17 cells, were lower in miR155 (−/−) Fas (lpr/lpr) than in Fas (lpr/lpr) mice. They concluded that miR155 might be a new target for therapeutic management in SLE (Reference Xin, Li and Dang96). However, an increment of miR155 expression was confirmed in PBMCs and CD19+ B cells, suggesting their use as diagnostic biomarkers for SLE patients (Reference Shumnalieva, Kachakova and Shoumnalieva-Ivanova97,Reference Stagakis, Bertsias and Verginis114) ; conversely, Lashine et al. detected lower levels in younger SLE patients. These contradictions could be explained by the heterogeneity of the disease and the differences observed in the pathological stages of this disorder (Reference Lashine, Salah and Aboelenein115). These authors correlated decreased expression of miR155 with IL2 deficiency associated with PP2Ac overexpression. Another miRNA implicated in the pathogenesis of SLE relating to IL2 impaired production is miR31. In fact, miR31 miRNA has been found to be markedly under-expressed in patients with SLE. It targets Ras homologue gene family member A (RhoA), a negative regulator of nuclear factor of activated T cells (NFAT) and cell apoptosis that increases the activity of IL2 (Reference Fan, Liang and Tang102). Therefore, under-expression of miR31 modulates a low production of IL2 through targeting RhoA in SLE T cells. Thus, deregulation of the miR31/RhoA axis represents a novel molecular mechanism that could counteract the IL2 deficiency in patients with SLE.
Emerging evidence has revealed an association between miRNA and DNA methylation. Some miRNAs such as miR21, miR148a and miR126 are overexpressed in CD4+ T cells from patients with SLE and contribute to inhibiting the expression of DNMT1 enzyme and inducing hypomethylation. Among these, miR126 modulates DNA methylation in CD4+ T cells from lupus patients and contributes to T- and B-cell autoreactivity in SLE by directly targeting DNMT1. As a consequence of hypomethylation on promoter region, an increase in CD11a and CD70 autoimmune-related protein levels is promoted (Reference Honarpisheh, Köhler and von Rauchhaupt12,Reference Zhao, Wang and Liang100) .
Besides, Stagakis et al. (2011) analysed miR21 in PBMCs from lupus patients. Compared with healthy CD4+ T cells, miR21 was significantly increased and correlated with SLE disease activity, showing enhanced IL10 production and CD40L expression through the suppression of programmed cell death protein (PDCD) 4, a selective protein inhibitor of genes involved in immune responses (Reference Stagakis, Bertsias and Verginis114).
In addition, miR142-3p/5p is involved significantly in pathogenesis of SLE (Reference Honarpisheh, Köhler and von Rauchhaupt12,Reference Gao, Liu and Min116) . Ding et al. (2012) speculated that in CD4+ T cells from SLE miR142-3p/5p is disrupted and acts to target signalling lymphocytic activation molecule-associated protein (SAP)-, CD84- and IL10- genes. It could be the cause of SAP, CD84 and IL10 increasing SLE-associated translation and T-cell and B-cell hyper-response (Reference Ding, Liang and Zhao101). Despite the fact that in lupus CD4+ T cells miR142-3p/5p intracellular levels are low, both were up-regulated in plasma and renal biopsies of SLE patients. This may be explained by exocytosis activity of these cells (Reference Stypinska and Paradowska-Gorycka117).
Decreased levels of miR1246, which target early B-cell factor (EBF) 1 mRNA specifically, have been found in B cells from SLE patients, inducing hyperactivation of B cells through surface co-stimulatory molecules CD40, CD80 and CD86. Activated B cells in SLE showed a positive feedback loop, where the lower expression of miR1246 via AKT-p53 signalling induces an up-regulation of EBF1 mRNA and B-cell activation (Reference Luo, Liu and Liang105). On the contrary, in CD4+ T cells from SLE, up-regulation of miR1246 could disrupt this amplifying mechanism. These findings provide a theoretical framework towards the research of novel biological targets in SLE treatment.
Several studies using microarray analysis detected other unusual miRNAs in PBMCs from patients with active lupus. They exhibited increased expression of certain miRNAs (miR189, miR61, miR78, miR342, miR299-3p, miR198, miR298, miR574-5p, miR1308, miR638, miR7) or decreased expression of others (miR196a, miR17-5p, miR409-3p, miR141, miR383, miR112, miR184, miR186, miR197), but thus far their role in the pathogenesis of disease is unknown (Reference Dai, Phillips and Zhang98,Reference Zhao, Wang and Liang100) .
lncRNAs act more specifically in biological functions than miRNAs, suggesting a strong implication in innate immunity and inflammatory response. lncRNAs can be measured in plasma, keeping unaltered in presence of RNAses; hence, they could be excellent biomarkers (Reference Wu, Li and Leng111). Besides, some lncRNAs allow for distinguishing patients with LN from SLE patients without kidney affection.
Genetic evidence suggests that gene encoding lncRNA growth arrest-specific (GAS) 5 is implicated in SLE susceptibility. GAS5 is required for the inhibition of human T-cell proliferation by rapamycin (Reference Zhao, Mao and Liu109). Several authors have measured lower levels of GAS5 in CD4+ T cells from SLE patients than from control subjects. However, the results did not show any remarkable differences regarding kidney affection between studied patients (Reference Suo, Sheng and Qiang118–Reference Shaker, Nassar and Gheta120).
Furthermore, abnormal expression of lncRNA nuclear paraspeckle assembly transcript (NEAT) 1 enhances the production of cytokines and chemokines in SLE patients, especially in monocytes where it regulates some genes (CXCL9, CXCL10, CXCL11, CCL8) related to IFN type I pathway. This fact could indicate that NEAT1 may facilitate the pathogenesis of SLE, and it may represent a novelty biomarker for diagnosis (Reference Zhao, Mao and Liu109,Reference Zhang, Wu and Qian110) .
Interestingly, some active patients of SLE disease present higher levels of linc0949, which is implicated in SLE disease activity index 2000 (SLEDAI-2K) score, nephritis and complement component C level and regulates IL6 and TNF-α production. linc0949 levels were decreased in PBMCs from lupus victims, so it could be used in monitoring of disease progression and treatment efficacy. Similarly, low expression of lnc0597 has been described in SLE PBMCs. However, it presents high levels in plasma. lnc0597 could be a regulator of IL6 and TNF-α cytokine expression, both of them implicated in SLE pathogenesis (Reference Wu, Li and Leng111).
Contrarily, metastasis-associated lung adenocarcinoma transcript (MALAT) 1, which regulates the sirtuin (SIRT) 1 signalling pathway and thus the production of IL21 in monocytes, was significantly increased in PBMCs and monocytes of lupus patients(Reference Zhao, Mao and Liu109). Considering that SLE patients exhibited higher levels of IL21 than healthy controls, we may conclude a critical role of MALAT1 in the SIRT1/IL21 pathway to lupus onset.
lnc-dendritic cell (lnc-DC) is implicated in the activation of STAT3 in dendritic cells and, subsequently, regulates T-cell activation, Th17 differentiation and IL2 production, participating in the pathogenesis of SLE. Significant differences in LN and SLE patients without nephritis have been shown. Thus, lnc-DC could be a useful biomarker to distinguish LN diagnosis in lupus patients (Reference Zhao, Mao and Liu109).
Epigenetic modifications as potential biomarkers and therapeutic targets
A comprehensive understanding of epigenetic approaches to autoimmune/inflammatory disease is necessary to predict individual disease outcomes and the introduction of effective, target-directed and tolerable therapies. Epigenetic events may, at least partially, explain inter-individual differences. In this line, epigenetic biomarkers are proving useful to verify molecular aberrations and environmental factors. Also, novel targets for individualised therapeutic interventions are making possible an advance in prevention, diagnosis and treatment. Nevertheless, epigenetic therapy needs further study.
Nowadays, drugs targeting epigenome alterations are focused on molecular inhibitors of DNMTs (DNMTi) and HDACs (HDACi).
In fact, several DNMTi have been developed, mainly as anti-cancer agents. These epigenetic modulators cause cell cycle and growth arrest, differentiation and apoptosis. Validation of co-therapies using these agents with cytotoxic drugs or radiotherapy could be crucial because the use of DNMTi offers improved access to cytotoxic agents or radiation for targeting DNA–protein complex. Therefore, there is a promising interest in the use of DNMTi as potential chemo- or radiation sensitisers to increase clinical response (Reference Gravina, Festuccia and Marampon121). Generically, DNMTi may be classified into two main classes, depending on their mode of action: nucleoside analogues and non-nucleoside analogues.
Firstly, nucleoside analogues consist of a cytosine ring connected to ribose or deoxyribose, which may integrate into DNA or RNA through replacement of cytosine units. Once incorporated into DNA and bonded to it covalently, they inhibit DNMTs activity (Reference Marques-Magalhães, Graça and Henrique122). To date, the most well-characterised nucleoside analogues are 5-azacytidine and 5′-aza-2′-deoxycytidine. Both of them were approved by the Food and Drug Administration (FDA) to treat myelodysplastic disorders (Reference Chistiakov, Orekhov and Bobryshev123), so most current preclinical and clinical trials evaluating functional effects and therapeutics targets are focused on cancer (Reference Marques-Magalhães, Graça and Henrique122,Reference Cuvillier, Schulz and Rollins124) . The treatment of human or mouse CD4+ T cells with these DNMTis induces or aggravates lupus-like disease, triggering T-cell autoreactivity and providing evidence of the implication of DNA methylation in autoimmunity (Reference Javierre and Richardson58). Richardson treated CD4+ T cells from healthy mice with 5-azacytidine, and then injected 5-azacytidine into identical animals. Mice receiving 5-azacytidine-treated cells expressed signs, anti-nuclear antibodies and complex glomerulonephritis closely related to human lupus (Reference Richardson125). Interestingly, several studies have demonstrated that the treatment of natural T cells with 5-azacytidine induced overexpression of different genes susceptible to methylation, as a consequence of DNMT inhibition. The genes identified were: the T–B cell co-stimulatory molecules CD70 and CD40L, the T-cell autoreactivity molecule CD11a, and the protein that integrates lethal pores into cell membrane perforin. All of them are hypomethylated and overexpressed in T cells from lupus patients (Reference Javierre and Richardson58,Reference Strickland and Richardson126–Reference Zhang, Lu and Chang128) . Treatment with 5-azacytidine has been used amply as drug-induced lupus, and it has enabled confirmation of the pattern of DNA methylation in SLE.
Additionally, other cytidine analogues have been developed to improve the properties of azanucleoside agents. Zebularine is a deoxycytidine derivative that lacks an amino group in position four of the pyrimidine ring (Reference Gowher and Jeltsch129). Its oral bioavailability and capacity to reactivate a silenced gene by oral administration have been well established (Reference Cheng, Yoo and Weisenberger130).
Another known DNMTi, 5-fluoro-2′-deoxycytidine, is a fluoropyrimidine nucleoside analogue that may bind covalently to DNMTs and form a suicide complex (Reference Mai and Altucci131); and 5,6-dihydro-5-azacytidine (DHAC) may be incorporated into RNA, inhibiting its synthesis and DNA methylation in human cell lines (Reference Cuvillier, Schulz and Rollins124). SGI-110 (guadecitabine) has been shown to be effective in DNA methylation inhibition in both in vitro and in vivo studies. Also, this second-generation drug seems to act as an immune modulator (Reference Srivastava, Paluch and Matsuzaki132). To improve 5-azacytidine, CP-400 was designed independently to the nucleoside transport system and showed more efficacy. Gemcitabine, the most-used treatment in cancer, is an anti-metabolite that acts with two active metabolites, inhibiting ribonucleoside reductase and/or in DNA replication supplanting a cytosine unit (Reference Marques-Magalhães, Graça and Henrique122).
Secondly, the non-nucleoside analogue group includes those compounds that bind to DNMTs covalently, without a previous interaction within DNA strand (Reference Marques-Magalhães, Graça and Henrique122). Some DNMTis of this group are procaine, procainamide, hydralazine, MG98 and RG108, among others. Procaine and procainamide act by inhibiting DNMTs, but they are also drugs usually employed as local anaesthetics and anti-arrhythmics, respectively. Nonetheless, procainamide has exhibited a more selective action on DNMT1 than on DNMT3a and DNMT3b (Reference Lee, Yegnasubramanian and Lin133). IM25 was derived from procainamide, within an improved toxic profile (Reference Marques-Magalhães, Graça and Henrique122).
Hydralazine was primarily used to treat high blood pressure. It forms strong hydrogen bonds with arginine and glutamic residues of DNMT1 (Reference Chistiakov, Orekhov and Bobryshev123). Like azanucleosides, most current studies about functional effects and therapeutics targets of non-nucleosides are focused on anti-cancer therapy. However, the treatment with procainamide or hydralazine disrupted genome-wide epigenetics and induced lupus-like disease. These drugs cause DNA hypomethylation in T cells and, thus, increased expression of LFA1, a molecule involved in T-cell activation, inducing autoreactivity. Similar murine studies have further confirmed that procainamide and hydralazine may induce overexpression of lupus-associated genes (CD70, CD40L, CD11a), considering them as SLE-induced drugs (Reference Javierre and Richardson58,Reference Strickland and Richardson126–Reference Zhang, Lu and Chang128) .
Complementarily, some non-nucleoside analogues exert precise DNMT inhibition. Nanaomycin A inhibits DNMT3b selectively; MG98, a second-generation short antisense nucleoside, acts on DNMT1 mRNA and down-regulates DNMT1 expression; and RG108 inhibits DNMT1 at catalytic domain. These second-generation drugs present a less cytotoxic profile (Reference Marques-Magalhães, Graça and Henrique122).
Other modifier drugs of DNA methylation target on DNMTs are methotrexate, an immune-suppressant used in rheumatic diseases, and cyclophosphamide, an antineoplastic drug (Reference Hedrich, Mabert and Rauen50). Methotrexate can deplete SAM levels, the substrate of DNMT, thus inhibiting it indirectly. On the other hand, cyclophosphamide induces DNMT1 activity, implementing DNA methylation. These epigenetic actions may provide a better understanding of the efficacy of treating SLE patients with methotrexate or cyclophosphamide (Reference Hedrich, Mabert and Rauen50).
The concept of precise modulation of methylated DNA in T-cell subsets has transformative potential in the treatment of ADs and beyond, but the global effects of DNMTis in different cell types render systemic treatment outcomes unpredictable, disturbing unaffected genes and deregulating them (Reference Li, Zhang and Zhong134). Unfortunately, SLE patients exhibit hypomethylation of genes, and only a few of them are hypermethylated; thus, the treatment with DNMTis reverting DNA hypomethylation is not an option.
Contrarily, different studies have reported HDACis improved autoimmune and inflammatory diseases (Reference Lewis, Blaabjerg and Storling135,Reference Grabiec, Krausz and de Jager137) . Accordingly, two HDACis have attracted special attention: trichostatin A (TSA) and suberoylanilide hydroxamic acid (SAHA or vorinostat), improving proteinuria, spleen weight and glomerulonephritis. Nanomolar concentrations of these have shown specific and reversible inhibition of HDACs. The effectiveness of TSA and SAHA on SLE disease has been tested in lupus murine models or in vitro, but no data are available yet about the restoration of histone acetylation and treatment in lupus patients. Daily TSA administration inhibited the production of inflammatory mediators in spleen and kidney of NBZ/W mice, leading to increased functionality of Treg cells associated with HDC inhibition (Reference Reilly, Thomas and Gogal138). In this line, Garcia et al. (2005) corroborated in MRL-lpr/lpr mice a correction of site-specific hypoacetylated after TSA treatment (Reference Garcia, Busby and Shabanowitz83). Mishra et al. (2003) reported the ability of TSA and SAHA in MRL-lpr/lpr mice. These inhibitors decreased levels of autoantibodies and proinflammatory cytokines such as IFN-γ, IL12, IL6 and IL10 mRNA, improving kidney and spleen affection. These inhibitors together were associated with HDACi efficacy to restore acetylation on H3 and H4 (Reference Mishra, Reilly and Brown139). Besides, these authors treated T cells from SLE patients with TSA, and significant down-regulation of CD154 and IL10 and up-regulation of IFN-γ were observed. Consequently, the capacity of TSA to interfere in inflammatory response through regulation of the CD40–CD154 axis (related to diverse immune system pathways), IL10 and IFN-γ expression supports its potential as a therapeutic agent to SLE (Reference Mishra, Brown and Olorenshaw140).
ACY-738, a specific HDAC6 inhibitor, increased splenic Tregs and decreased Th17 cells in NZB/W mice, related to decreased serum anti-dsDNA levels. Moreover, ACY-738 improved the phenotype of SLE by inhibiting the production of IL6, IL10 and TGF-β, and deposition of C3 and IgG in glomeruli, reducing LN (Reference Mishra, Brown and Olorenshaw140). CKD-506, another inhibitor of HDAC6, enhanced survival rate and decreased proteinuria, kidney inflammation and glomerular infiltration of IgG and C3 in NZB/W F1 mice. Furthermore, CKD-506 reduced levels of inflammatory cytokines in serum and kidney (Reference Choi, Song and Ha142). Recent studies showed that ITF2357 (givinostat), an inhibitor of class I and II HDACs that is used mainly in juvenile idiopathic arthritis, increased Treg cells and Foxp3 acetylation and, thus, inhibited autoantibody and pro-inflammatory cytokine production in NZB/W mice. Experiments in animals showed SLE reduced serum and urine biomarkers (Reference Regna, Chafin and Hammond143).
Based on these studies, it has been suggested that HDACi may be a promising new therapy for SLE. Nevertheless, the clinical effects of HDACi in lupus patients have not been demonstrated yet. Conversely, histone-modifying drugs have been used for epilepsy (valproic acid) and oncology (vorinostat, romidepsin) therapies (Reference Hedrich6,Reference Burke, Lamba and Pound144) . Additionally, the FDA approved vorinostat for the treatment of cutaneous T-cell lymphoma in 2006 (Reference Duvic and Vu145,Reference Störman and Schophl146) , used in monotherapy or in combination with other drugs (Reference Iwamoto, Friedam and Sandhu147–Reference Pinto, DuBois and Marachelian149). On the other hand, givinostat improved juvenile idiopathic arthritis (Reference Vojinovic, Damjanov and D’Urzo150) and reduced muscle fibrosis in Duchenne muscular dystrophy, showing an excellent safety profile (Reference Bettica, Petrini and D’Oria151).
In short, epigenetic markers and immune-associated activity may be used as novel therapeutic targets and biomarkers, supplying possible alternatives to clinical treatment in SLE. Future research and investigations about epigenome in specific cell types and organs are needed to support the mechanism of action of these DNMTi and HDACi ‘epidrugs’ in the disease and their future applications in SLE. This aspect should be studied in depth, evaluating the efficacy of therapy within these agents, alone or in combination with existing therapies in clinical trials of patients with lupus disorders, which complement the results obtained in recently published preclinical studies.
Diet, epigenetic and SLE linkage
The pathogenic process of ADs is considered a complicated and complex challenging question. Genetic predisposition and environmental factors, such as nutrition, infection, pollution or UV exposure, can cause autoimmunity and are implicated in the pathogenesis, although how much of each is not well known yet. Nutrition and dietary habits are one of the most accessible external factors, with diet being a possible tool to act on the disease onset. Generally, it has been recognised that feeding habits, such as calorie restriction or supplementation with macronutrients (fibre, unsaturated fatty acids or specific bioactive compounds (Reference Nobs, Zmora and Elinav152)), may influence development and predisposition of pathologies, especially ADs (Reference Szarc, Ndlovu and Haegeman153). A growing number preclinical and clinical studies have evidenced the relation of dietary components and patterns with the immune response (Reference Venter, Eyerich and Sarin154). A clear example of this is the discordance in the incidence of immune-mediated diseases in Western countries. In fact, the Mediterranean diet might support an improvement in diseases related to the immune system, inflammation or oxidative stress, confirmed in RA, SLE, cardiovascular disease (CVD), cancer and neurodegenerative disease patients, among others (Reference Barrea, Muscogiuri and Frias-Toral155–Reference Casas, Sacanella and Estruch159). This diet emphasises vegetables, fruits, nuts, fish and olive oil (OO) as a main fat source.
Focusing on SLE disorders, there has been a growing body of preclinical and clinical literature describing the control of lupus disease by dietary interventions (Reference La Cava160). A high-quality diet is of notable importance since these patients used to be affected with vitamin and mineral deficiencies, anaemia or high CVD susceptibility (Reference Aparicio-Soto, Sánchez-Hidalgo and Alarcón-de-la-Lastra10). A balanced diet and its nutrients exhibit antioxidant, anti-inflammatory and immunomodulatory properties (Reference Aparicio-Soto, Sánchez-Hidalgo and Alarcón-de-la-Lastra10). Accordingly, a nutritional therapy based on carbohydrate and protein restriction, and vitamin (A, B, C, D and E) and omega-3 supplementation with adequate fibre and sodium consumption, enables a reduction in the severity, or prevention, of disease (Reference Aparicio-Soto, Sánchez-Hidalgo and Alarcón-de-la-Lastra10,Reference Venter, Eyerich and Sarin154,Reference Estruch158–Reference de Medeiros, Medeiros and de Medeiros162) . In spite of multiple studies suggesting the beneficial properties of nutritional therapy (Reference Islam, Khandker and Kotyla161,Reference de Medeiros, Medeiros and de Medeiros162) , more clinical trials are needed and further research is required.
In this respect, a cross-relation between ADs, diet and epigenetics may exist. Several authors have shown that certain early diet habits may lead to metabolic and physiology changes through epigenetic altered pattern, defining susceptibility to chronic pathologies with the passage of time (Reference Szarc, Ndlovu and Haegeman153,Reference Mckay and Mathers163,Reference Franzago, Santurbano and Vitacolonna164) . Based on diet restriction and following specific eating habits, such as Mediterranean diet or consumption of certain nutrients, they have shown their implication for modulation of DNA methylation, histone patterns or ncRNA expression (Reference Mckay and Mathers163,Reference Corella, Coltell, Macian and Ordovás165–Reference Ng, Fann and Jo168) . Even following specific pregnancy-feeding guidelines seems to play a key role in the epigenetic profile of embryo (Reference Szarc, Ndlovu and Haegeman153).
Diet is an essential player in homeostasis of epigenetic marks, making it a top influencer for SLE development (Reference Somers and Richardson127). To date, there are some studies about nutrients’ effect on HDAC and HAT activity or miRNA expression, but diet’s influence on DNA methylation is better established. There are two main mechanisms that alter DNA methylation: (i) availability of methyl donors, and (ii) disruption of DNMT1 activity (Reference Mckay and Mathers163). The most widely investigated of them is the influence of nutrients on methyl group supply for one-carbon metabolism.
Partly, this may be due to levels of SAM, a methyl donor, dependent on dietary micronutrients, such as folate, zinc, methionine, choline, and B6 and B12 vitamins, and oxidative stressors and age, which may decrease DNMT1 activity. In SLE patients, high oxidative response and sensitivity to diets deficient in SAM donors has been observed. For this reason, the maintenance of balance between DNMT1 and SAM levels is a key point to avoid SLE flares and onset (Reference Somers and Richardson127,Reference Li, Liu and Strickland169) . Accordingly, Strickland and colleagues observed that dietary micronutrients which are implicated in methylation process may affect genes through epigenetic mechanisms in correlation with the severity of disease (Reference Strickland, Hewagama and Wu170). A transgenic lupus model with inducible ERK pathway signalling defect was bred onto lupus-resistant (C57BL/6) and lupus-susceptible (C57BL/6xSJL) mouse strains. These mice express a dominant-negative MEK uniquely in CD2+ cells when doxycycline, a DNMT1 level regulator, is administered in their drinking-water. Both mouse groups that were fed a diet with low levels of methyl-related nutrients (choline and methionine) showed high renal damage and developed haematuria, while mice fed with a diet supplemented with high levels of methyl donors reduced the development of kidney disease. Taken together, the authors concluded that dietary micronutrients can contribute to lupus susceptibility and severity through genetic/epigenetic interactions that affect DNA methylation.
Additionally, SLE patients present decreased levels of vitamins, methionine and other methyl-donors in comparison with healthy individuals. Nevertheless, diet supplemented with vitamins (A, B6, C, D or E) resulted in improvement of flares and symptoms of SLE (Reference Aparicio-Soto, Sánchez-Hidalgo and Alarcón-de-la-Lastra10). Ray et al. (2018) hypothesised that expression of genes susceptible to DNA methylation in T cells from SLE patients may be more prevalent in low-micronutrient conditions than in healthy T cells. They cultured CD4+ T cells from lupus patients PHA-stimulated in culture media with normal or low levels of B6 and B12 vitamins, methionine, folate and choline, and then measured expression of CD70, perforin and KIR (Reference Ray, Strickland and Richardson171). It should be noted that KIR, perforin and CD70 genes are usually hypermethylated and overexpressed in SLE (Reference Javierre and Richardson58,Reference Richardson125–Reference Zhang, Lu and Chang128) . The authors observed that CD4+ T cells from SLE patients cultured in low levels of supplements exhibited an increase in these methylation-sensitive genes (Reference Ray, Strickland and Richardson171). In conclusion, SLE patients may pay attention to their nutrition to avoid low levels of methyl donors and DNMT activity, preventing lupus flares and onsets, especially older people with more diet deficiencies.
A large number of articles have reported independently that some bioactive compounds consumed in diet exert beneficial properties on SLE, and these same micronutrients act via epigenetic modulation (Reference Aparicio-Soto, Sánchez-Hidalgo and Alarcón-de-la-Lastra10,Reference Evans, Stratton and Ferguson172,Reference Hardy and Tollefsbol173) , although most of them have been established in cancer. Taking these properties into consideration, we could analyse these studies in order to clarify the cross-relation among the consumption of these nutraceuticals, the improvement in SLE disease profile and their effects on epigenetic machinery (Table 4). Some of these dietary bioactive compounds could be curcumin, epigallocatechin gallate (EGCG), resveratrol, genistein, indole-3-carbinol (I3C) or OO.
Table 4. Overview of micronutrients related to SLE pathological and epigenetic effects

Curcumin, a polyphenol from the golden spice turmeric, inhibited cell proliferation and differentiation and ameliorated the immune response of Th1 through IFN-γ inhibition. Besides, this polyphenol reduced Th17 cell response, inhibiting the expression of relevant proinflammatory cytokines, and modulated Th17/Treg cell balance in CD4+ from SLE patients, exhibiting therapeutic profile to this AD (Reference Boyanapalli and Kong174–Reference Momtazi-Borojeni, Haftcheshmeh and Esmaeili176). In experimental animal studies, curcumin-supplemented diet ameliorated renal damage and controlled autoantibody and cytokine production on lupus onset Reference Wu, Chang and Lu66,Reference Momtazi-Borojeni, Haftcheshmeh and Esmaeili176,Reference Dent, Taylor and Turbeville177) . Similar effects were observed in clinical trials using curcumin-supplemented diet or curcumin-treated T cells from SLE patients, regulating Th17/Treg balance of CD4+ T cells (Reference Aparicio-Soto, Sánchez-Hidalgo and Alarcón-de-la-Lastra10,Reference Dent, Taylor and Turbeville177,Reference Khajehdehi, Zanjaninejad and Aflaki178) . Complementarily, curcumin is known to decrease HAT activity at H3ac of IL6 promoter, inhibiting the release of IL6 and TNF-α (Reference Kumari, Bhawal and Kapila180). Balasubramanyam and colleagues (2004) postulated that curcumin was the first known p300 specific natural HAT inhibitor in transcription (Reference Balasubramanyam, Varier and Altaf181). Moreover, daily intake of curcumin decreased miR21 and miR155 levels and suppressed DNMT1, DNMT3a and DNMT3b expressions (Reference Kumari, Bhawal and Kapila180).
In this line, different in vitro and in vivo studies support the modulation action of the green-tea polyphenol ECGC on NF-kB, nucleoside binding domain (NOD)-like receptors (NLRP) 3, nuclear factor erythroid-derived (Nrf) 2 and T-cell activation. Overall, EGCG controls inflammation and prevents renal function impairment. In this line, several in vivo studies showed depletion of progression in lupus-like syndrome, including glomerulonephritis and autoantibody production in ECGC-fed mice (Reference Aparicio-Soto, Sánchez-Hidalgo and Alarcón-de-la-Lastra10,Reference Tsai, Ka and Chang182–Reference Sayama, Oguni and Tsubura184) . With regard to epigenetics, EGCG decreased HDACs and HAT class I activity that affected NF-kB, IL6 expression and inflammatory response (Reference Thakur, Gupta and Gupta185), but also, EGCG inhibited histone methyltransferase EZH2, which adds a methyl group to H3K27. Moreover, this green-tea polyphenol inhibited DNA methylation through an interaction with DNMT1. It was able to form four hydrogen bounds on the catalytic centre of DNMT1, rejecting the entrance of cytosine residue (Reference Fang, Wang and Ai186). Also, DNMT3a and DNMT3b were inhibited in an in vivo study with EGCG-treated mice (Reference Wong, Nguyen and Noh187). Complementarily, antioxidant properties of green tea could be explained by restoring expression of antioxidant enzymes, as glutathione S-transferase P (GSTP) 1 by inducing demethylation and DNMT inhibition (Reference Chistiakov, Orekhov and Bobryshev123).
Resveratrol, a polyphenol present in tomato, peanuts or skin of red grapes, showed protective effects in lupus disorders, and supported its ability to regulate inflammatory genes and transcription factors such as STAT3, NF-kB or cyclooxygenase (COX) 2, implicated in SLE disease (Reference Nakata, Takahashi and Inoue188,Reference Wang, Luo and Li189) . Consequently, in oxidative stress and inflammatory response, resveratrol treatment decreased the expression and activity of DNMT3a and SIRT1, ameliorating harmful conditions (Reference Roy, Chen and Fu190). In addition, resveratrol treatment in human THP-1 macrophages restored levels of pro-inflammatory cytokines through miR-Let7A overexpression (Reference Song, Jun and Ahn191), and controlled miR21 levels, down-regulating NF-kB, TNF- α, IL1β and IL6 (Reference Chistiakov, Orekhov and Bobryshev123).
I3C is abundant in cruciferous vegetable such as broccoli, cabbage or cauliflower. In NZB/w F1 mice fed with a I3C-supplemented diet, proteinuria and renal affections were decreased and survival of animals was increased (Reference Auborn, Qi and Yan192). These data confirmed a beneficial effect of dietary I3C in experimental SLE.
Recently, Eghbalpour et al. (2020) investigated the effect of I3C on transcriptional profiling of macrophage-derived monocytes (MDMs) from SLE patients in four stages of the wound-healing process. The results showed that treatment with I3C could modulate STAT1, THBS1 and ATP2A3 gene expression involved in wound healing in SLE cases and healthy controls (Reference Eghbalpour, Aghaei and Ebrahimi193). Additionally, I3C counteracted the effects of staphylococcal enterotoxin B (SEB), a superantigen inductor of inflammation and immune cell activation, in SEB-stimulated T cells. This indole re-established pro-inflammatory cytokines levels and cellular infiltration, inhibited HDAC class I and decreased miR31 production. Busbee and colleagues suggested that the regulatory epigenetic action associated with I3C was the reason for its immune-inflammatory regulation capacity (Reference Busbee, Nagarkatti and Nagarkatti194,Reference Busbee, Nagarkatti and Nagarkatti195) .
The isoflavone genistein that is present in soybeans is well known for its antioxidant effects. Genistein improved the severity of disease and regulated autoantibody production, proteinuria and renal pathology in lupus patients (Reference Aparicio-Soto, Sánchez-Hidalgo and Alarcón-de-la-Lastra10). Recently, Li et al. (2019) reported that genistein may recover epigenetic aberrations of Klotho (kidney anti-aging and fibrosis-suppressing protein) in a mouse kidney disease incurred by unilateral ureteral occlusion. The authors used genistein-treated human kidney tubular cells (HK2) and C57BL/6 mouse kidney lysates. Genistein inhibited DNMT1, DNMT3a and DNMT3b and also decreased histone acetylation, alleviating renal fibrosis by Klotho restoration (Reference Li, Chen and Wei196). Additionally, levels of miR155 were down-regulated in MDA-MB-435 and Hs578t cancer cells treated with genistein (Reference De la Parra, Castillo-Pichardo and Cruz-Collazo197). The potential effectiveness of genistein in the restoration of epigenome alterations suggests it is an interesting dietary supplement in prophylaxis and treatment of several ADs and cancer.
The beneficial properties of OO in SLE have been described widely, particularly extra virgin olive oil (EVOO). Aparicio-Soto and colleagues have demonstrated that SLE-induced mice fed with EVOO-supplemented diet presented lower renal incidence and inflammatory mediators, such as matrix metalloprotease (MMP) 3, prostaglandin (PG) E2, pro-inflammatory cytokines, Nrf-2/haem oxygenase (HO) 1, MAPKs and NF-kB signalling pathways (Reference Aparicio-Soto, Sánchez-Hidalgo and Cárdeno198). These authors also reported that polyphenolic fraction of EVOO modulated cytokine production, attenuating T-cell activation using PBMC from lupus patients (Reference Aparicio-Soto, Sánchez-Hidalgo and Cárdeno199). In terms of DNA methylation, EVOO induced changes in the expression of several inflammatory-related genes in peripheral leucocytes from patients fed a Mediterranean diet (Reference Arpón, Milagro and Razquin200). Moreover, OO exerted demethylating activity and inhibited DNMT1 (Reference Nanda, Mahmood and Bhatia201).
In summary, dietary interventions could modify the epigenetic profile, with numerous studies consolidating this affirmation (Fig. 2). Although several publications report correlating changes in epigenetic-specific alteration in some genes and its expression in response to dietary bioactive compounds, to date, few of them have investigated both influence on lupus and the functional consequences of modifying nutrition habits.
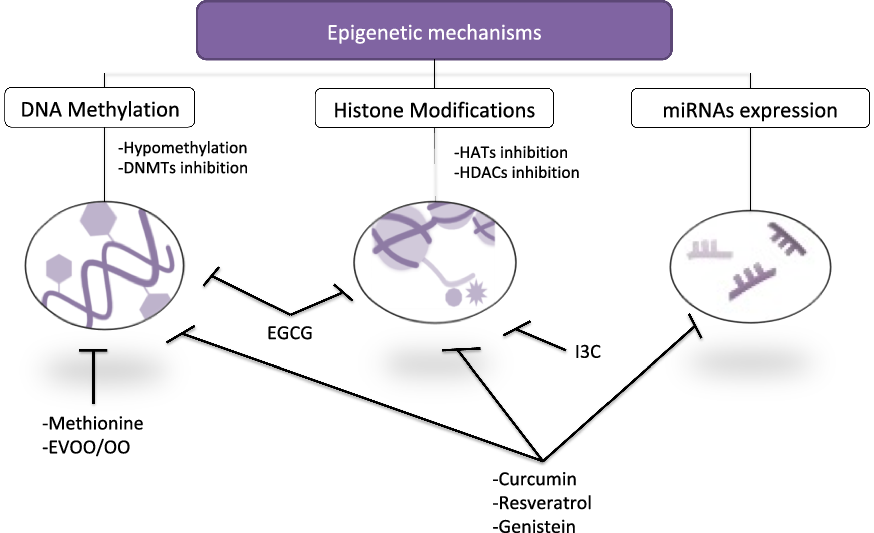
Fig. 2. Schematic representation linking key epigenetic mechanism and bioactive compounds consumed in diet. Several micronutrients exhibited regulatory properties on epigenetics through hypomethylation of DNA, DNMT inhibition and/or regulation of HATs, HDACs and miRNAs. EGCG, epigallocatechin gallate; EVOO/OO, extra virgin olive oil/olive oil; I3C, indole-3-carbinol.
Conclusion
There is growing evidence for the implication of environmental factors, such as nutrition, lifestyle or UV exposure, that orchestrate epigenetic changes in predisposed subjects, contributing to modulation of SLE onset. Generally, global losses of DNA methylation and H3 and H4 histone deacetylation have been identified in lupus disorder. Despite the fact that more accurate and revealing preclinical and clinical works are required for understanding the pathogenic mechanism contributing to SLE, epigenome patterns have been proposed as novel approaches to SLE therapy.
Accordingly, DNA methylation and histone modification have been postulated as key targets that could be employed in the development of individual and precise therapies with lower or no side effects. Particularly, recent studies have suggested the testing of some ‘epidrugs’, whose mechanism of action is based on DNMT or HDAC inhibition, underscoring the capacity of these drugs to modulate target gene expression defined in SLE.
In fact, the measurement of ncRNAs levels in serum or certain organs has been established as useful diagnosis tools. Indeed, significant differences in several miRNAs’ levels were observed between lupus patients and normal controls, or even in lupus patients with kidney affection compared with those without kidney damage. These revealing facts suggest miRNAs as new strategies for diagnosis and therapeutic targets of lupus.
Improvement of disease associated with some micronutrients has been widely demonstrated; notwithstanding, little is known about their relation with epigenetic alteration in SLE. Although studies are not conclusive and further research is required, it seems to be clear that diet supplemented with certain bioactive compounds could be effective in preventing or avoiding aberrant modifications in epigenome to improve SLE onset. Therefore, the combined use of epigenetic bioactive compounds with therapeutic drugs could be of interest because of the possible synergistic effects that may be observed in the treatment of disease.
Mainly dietary bioactive compounds have been studied within cancer mice models and only a few preclinical trials, so emergence studies in SLE and immune-inflammatory disorders are required for a better understanding. Nonetheless, the effects of the dietary bioactive compound on the epigenome, a so-called epigenetic diet, could offer an approach in prevention, therapy and diagnosis of diseases in the future.
Financial Support
The authors acknowledge the financial support by the research grant from the Ministerio de Economía y Competitividad of Spain (AG2017-89342-P). T.M. thanks the Postgraduate Program of PIF fellowship and financial sponsorship from VI Plan Propio de Investigación y Transferencia de la Universidad de Sevilla. R.M. gratefully acknowledges the FPU fellowship from Ministerio de Educación y Cultura of Spain.
Conflicts of Interest
The authors declare no conflict of interest.
Authorship
C.A.-d.-l.-L. and T.M. conceived the idea. M.L.C., R.M.-G., T.M. and C.A.-d.-l.-L. reviewed the published literature and wrote the first draft of the manuscript. C.A.-d.-l.-L. and T.M. revised and edited the final manuscript. All authors contributed and approved the published version of the manuscript.









