1. Introduction
With the rapid development of health information systems, more and more electronic health records (EHRs), such as clinical narratives and discharge summaries, are available for research (Figure 1). Extracting clinical entities like disorders, drugs, and treatments from EHRs has become a topic of increasing interest (Figure 2). This task is important because it can help people understand the potential causes of various symptoms and build many useful applications for clinical decision support systems. Moreover, the extraction of these entities forms the basis for more complex tasks, for example, entity linking, relation extraction, and document retrieval.
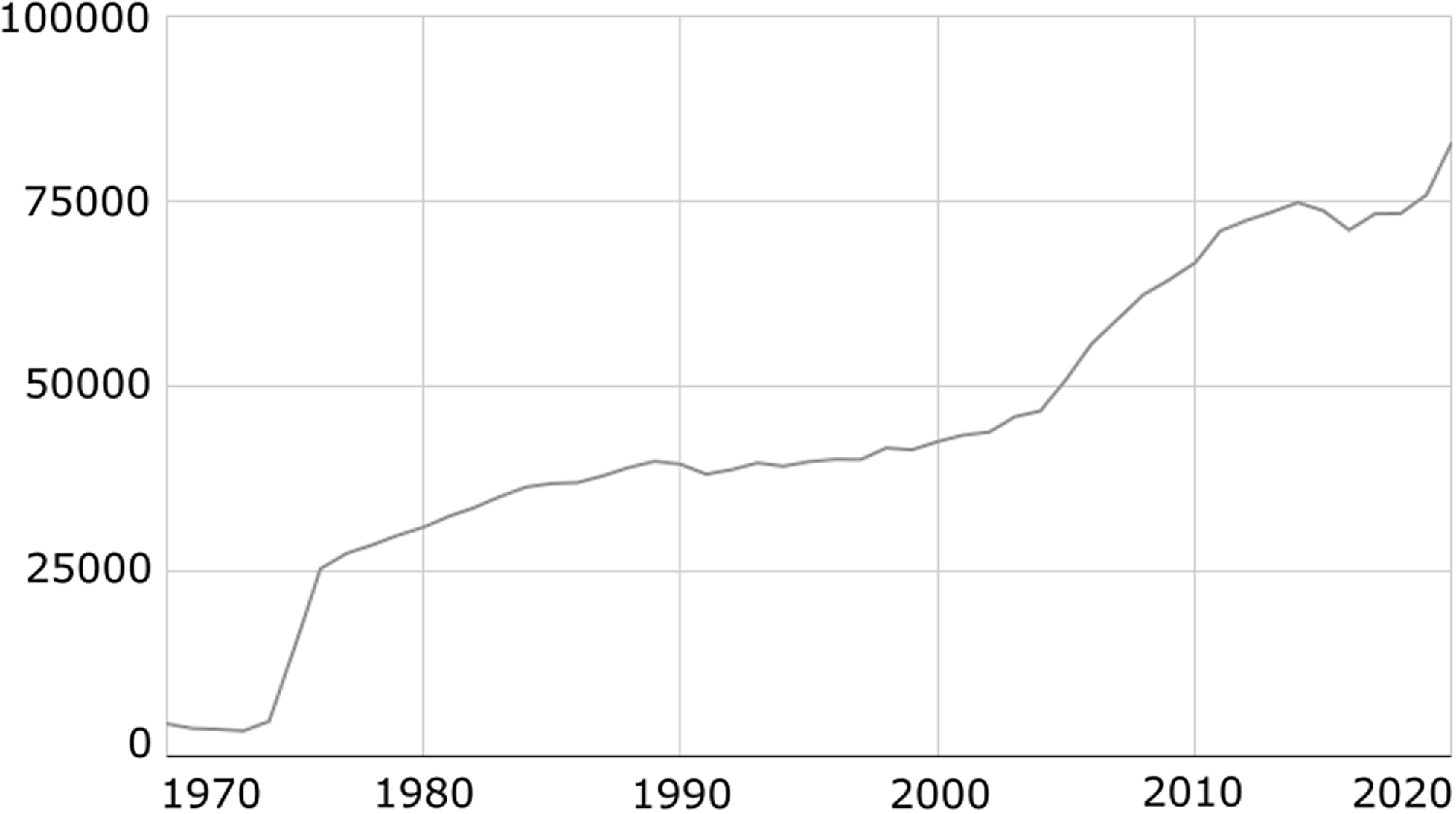
Figure 1. Number of published case reports per year in PubMed extracted with the query “Case Reports[Publication Type]”.

Figure 2. Number of publications per year in PubMed about entity recognition extracted with the query “Entity Recognition[Title/Abstract]”.
Since EHRs have an unstructured format, the entities of interest must first be identified and extracted before being queried and analyzed.
Disorder named entity recognition (DNER) is the natural language processing (NLP) task of automatically recognizing named entities of disorders in medical documents. For instance, the excerpt below contains four disorder entities, that is, “abdominal pain”,“fever”,“fatigue”,and “CML”.

DNER is considered a challenging problem, mainly due to name variations of entities. In fact, disorder entities can appear in the text in many forms that are different from their standard names. For example, the entity diplopia can be mentioned in the text as seeing double images. Moreover, entities are often ambiguous and context dependent, for example,“stroke” is a disorder in “compatible with an acute ischemic stroke”; however, it does not refer to the disorder in “to increase the stroke volume with further fluids”. Entities can also consist of long multi-word expressions (e.g., “lesion in the mid portion of the left anterior descending coronary artery”) that make the task of DNER even more difficult.
While it is true that existing tools for DNER have traditionally relied on rule-based and dictionary lookup methods (e.g., MetaMap (Aronson Reference Aronson2001) and cTAKES (Savova et al. Reference Savova, Masanz, Ogren, Zheng, Sohn, Kipper-Schuler and Chute2010)), the recent advancements in deep learning methods, such as BERT, have reshaped the field. Models like PubMedBERT (Gu et al. Reference Gu, Tinn, Cheng, Lucas, Usuyama, Liu, Naumann, Gao and Poon2021), CancerBERT (Zhou et al. Reference Zhou, Wang, Wang, Liu and Zhang2022), and HunFlair (Weber et al. Reference Weber, Sänger, Münchmeyer, Habibi, Leser and Akbik2021) have gained prominence and demonstrated remarkable performance in DNER tasks.
European Clinical Case Corpus (E3C) (Magnini et al. 2020, 2021) is a freely available multilingual corpus (English, French, Italian, Spanish, and Basque) of semantically annotated clinical narratives that has been recently made available to the research community. In the corpus, a clinical narrative is a detailed report of the symptoms, signs, diagnosis, treatment, and follow-up of an individual patient, as illustrated in the extract below.

The clinical narratives in the E3C corpus were collected from both publications, such as PubMed, and existing corpora like the SPACCC corpus, and also from admission tests for specialties in medicine.
The E3C corpus consists of two types of annotations:
• clinical entities (disorders), which are annotated at the mention and concept level. The mention-level annotation contains the entity text spans covering disorder entities, for example, renal colic. The concept-level annotation was obtained by linking the annotated entities to their corresponding concepts in the Unified Medical Language System (UMLS) (Bodenreider Reference Bodenreider2004), for example, C0156129.
• temporal information, including events, time expressions, and temporal relations according to the THYME standard (Styler IV et al. Reference Styler, Bethard, Finan, Palmer, Pradhan, de Groen, Erickson, Miller, Lin, Savova and Pustejovsky2014).
In this article, a gap is filled concerning the exploitation of the E3C corpus for the development of information extraction systems, showing the appropriateness of the annotation of clinical entities for DNER tasks.
In this study, the clinical entity annotation of the E3C corpus has been used to train machine learning (ML) models for DNER. Specifically, the mention-level annotation has been exploited to compare traditional ML models like conditional random fields (CRFs) (Lafferty, McCallum, and Pereira Reference Lafferty, McCallum and Pereira2001) with more recent multilingual pre-trained models like XLM-RoBERTa (Conneau et al. Reference Conneau, Khandelwal, Goyal, Chaudhary, Wenzek, Guzmán, Grave, Ott, Zettlemoyer and Stoyanov2020). Concerning the multilingual pre-trained models, they were evaluated on each language of the corpus by using three different configurations: (i) training on data in the target language (monolingual training and evaluation), (ii) training on data in all languages (multilingual training and evaluation), and (iii) training on data in all languages except for the target language (cross-lingual training and evaluation).
The results obtained with the above models were compared with the results of two baselines: (i) the CoNLL-2003 baseline (Tjong Kim Sang and De Meulder Reference Tjong Kim Sang and De Meulder2003), which only recognizes entities that appear in training data and (ii) a dictionary lookup baseline that uses the disorder entities listed in UMLS to find the relevant entities in the text. In this comparison, the proposed models outperformed the CoNLL-2003 baseline and were competitive with the dictionary lookup baseline. The considered models performed also in line with the models that took part in (i) Task 1 of the ShARe/CLEF eHealth Evaluation Lab 2013 (Mowery et al. Reference Mowery, Velupillai, South, Christensen, Martínez, Kelly, Goeuriot, Elhadad, Pradhan, Savova and Chapman2013), which focused on recognition of disorder entities in clinical reports written in English and (ii) Task 3 of the DEFT 2020 challenge (Cardon et al. Reference Cardon, Grabar, Grouin and Hamon2020), which required the recognition of clinical entities in a corpus of clinical cases in French.
Once the entities have been recognized, they can be linked to concepts in standard vocabularies such as UMLS. To show the appropriateness of the concept-level annotation of the corpus for entity linking tasks, a preliminary experiment has been conducted in which a basic dictionary lookup method has been tested in cascade to the considered models for DNER.
The findings of the present work show the appropriateness of the E3C corpus for training ML models for DNER. The trained models can also be used to annotate clinical case texts in languages other than the languages of the corpus. Researchers can use these results as baselines for this corpus against which to compare their results. The top ranked model resulting from this study has been made available through the European Language Grid (ELG) platform (Rehm et al. Reference Rehm, Piperidis, Bontcheva, Hajic, Arranz, Vasiļjevs, Backfried, Gomez-Perez, Germann, Calizzano, Feldhus, Hegele, Kintzel, Marheinecke, Moreno-Schneider, Galanis, Labropoulou, Deligiannis, Gkirtzou, Kolovou, Gkoumas, Voukoutis, Roberts, Hamrlova, Varis, Kacena, Choukri, Mapelli, Rigault, Melnika, Janosik, Prinz, Garcia-Silva, Berrio, Klejch and Renals2021). This allows for easy access to experiment with it.
This work is structured as follows. Section 2 briefly surveys related work. Section 3 describes the E3C corpus. Section 4 provides details on the procedure followed to conduct the experiments of this work. Section 5 shows the results obtained by the models. Finally, Section 6 presents and discusses the overall results.
2. Related work
Over the past decade, biomedical named entity recognition (BNER) has acquired more and more relevance, with the ever-increasing availability of biomedical documents and the corresponding deluge of biomedical entities scattered across them. As a matter of fact, the very unstructured and chaotic nature of biomedical literature, with little to no compliance with agreed-upon standards or naming conventions, colliding or polysemous acronyms and terms, etc., has driven researchers to try and develop appropriate mechanisms to retrieve structured information from it as automatically as possible. These mechanisms ranged from rule-based systems, statistical NLP, up to ML-based approaches, the earliest of which appearing in literature more than 10 years ago for a number of purposes (as in Atzeni, Polticelli, and Toti (Reference Atzeni, Polticelli and Toti2011); Toti, Atzeni, and Polticelli (Reference Toti, Atzeni and Polticelli2012), for instance).
In the clinical domain, BNER has gained even more attention since some annotated datasets have become available in challenges such as ShARe/CLEF eHealth Evaluation Lab 2013 (Mowery et al. Reference Mowery, Velupillai, South, Christensen, Martínez, Kelly, Goeuriot, Elhadad, Pradhan, Savova and Chapman2013), BC5CDR (Li et al. Reference Li, Sun, Johnson, Sciaky, Wei, Leaman, Davis, Mattingly, Wiegers and lu2016), and n2c2 (Henry et al. Reference Henry, Buchan, Filannino, Stubbs and Uzuner2019). In relation to annotated datasets for languages other than English, it is worth mentioning the CAS corpus of clinical cases annotated in French (Grabar, Dalloux, and Claveau Reference Grabar, Dalloux and Claveau2020) and the multilingual E3C corpus (Magnini et al. 2020, 2021), which is the subject of this study.
Many ML models for BNER are models that have been widely used for entity recognition in the newswire domain. Among these models, CRF (McCallum and Li Reference McCallum and Li2003) was the most commonly used.
More recently, deep learning models have been demonstrated to be very effective in many tasks of NLP, including named entity recognition (NER). Long short-term memory (LSTM) combined with CRF has greatly improved performance in BNER (Giorgi and Bader Reference Giorgi and Bader2019). Word representation models such as Word2Vec (Mikolov et al. Reference Mikolov, Chen, Corrado and Dean2013) have become popular as they can improve the accuracy of ML methods. With these models, words with similar meaning have a similar representation in vector format. For example, fever and pyrexia are closer in distance (and hence more semantically similar) than words with completely different meanings like fever and muscular pain. Lample et al. (Reference Lample, Ballesteros, Subramanian, Kawakami and Dyer2016) combined the power of word vector representation models, LSTMs and CRF, into a single method for entity extraction. One of the limitations of word representation models like Word2Vec is that they produce a single vector representation for each word in the documents, ignoring the context where the word appears. Unlike Word2Vec, BERT (Devlin et al. Reference Devlin, Chang, Lee and Toutanova2019) considers the context word order and learns different representations for polysemous words. In their study, the authors of BERT showed that pre-training such a contextual representation from large unlabeled texts, followed by fine-tuning, achieves good performance even when labeled data are scarce. RoBERTa (Liu et al. Reference Liu, Ott, Goyal, Du, Joshi, Chen, Levy, Lewis, Zettlemoyer and Stoyanov2019) modified some key hyper-parameters of BERT and trained on much larger amounts of data. BioBERT (Lee et al. Reference Lee, Yoon, Kim, Kim, Kim, So and Kang2019) demonstrated that pre-training BERT on additional biomedical corpora helps it analyze complex biomedical texts.
Finally, multilingual transformer models like mBERT (Devlin et al. Reference Devlin, Chang, Lee and Toutanova2019) and XLM-RoBERTa (Conneau et al. Reference Conneau, Khandelwal, Goyal, Chaudhary, Wenzek, Guzmán, Grave, Ott, Zettlemoyer and Stoyanov2020) have obtained great improvements for many NLP tasks in a variety of languages. Specifically, XLM-RoBERTa is a large multilingual language model that was trained on 2.5 TB of data across 100 languages, including the five languages of the E3C corpus. These multilingual models enable to train and evaluate per-language data or perform cross-lingual learning by training on one language data and evaluating on another different language data.
Many popular tools for BNER are based on dictionary lookup methods. For example, MedLEE (Friedman Reference Friedman2000), MetaMap (Aronson Reference Aronson2001), and cTAKES (Savova et al. Reference Savova, Masanz, Ogren, Zheng, Sohn, Kipper-Schuler and Chute2010). With most recent tools, such as DNORM (Leaman, Islamaj Doğan, and Lu ), NER is initially performed using ML-based methods which is followed by entity linking that can be rule- or ML-based. One of the main drawbacks of this cascade approach is that it suffers from error propagation, an inherent drawback of any pipeline architecture. To overcome this issue, OGER (Furrer, Cornelius, and Rinaldi Reference Furrer, Cornelius and Rinaldi2020) uses a parallel architecture, where NER and entity linking are tackled in parallel.
New state-of-the-art BNER tools such as HunFlair (Weber et al. Reference Weber, Sänger, Münchmeyer, Habibi, Leser and Akbik2021) and BERN2 (Sung et al. Reference Sung, Jeong, Choi, Kim, Lee and Kang2022) are based on deep neural networks. The training of HunFlair is a two-step process. First, in-domain word embeddings are trained on a large unlabeled corpus of biomedical articles, which are then used in the training of the NER tagger on multiple manually labeled NER corpora. BERN2 uses a multi-task NER model to extract biomedical entities, followed by a neural network-based model to normalize the extracted entities to their corresponding entity identifiers in MESH. An overview of the main deep learning methods used in BNER is presented in Song et al. (Reference Song, Li, Liu and Zeng2021).
3. The E3C corpus
The E3C multilingual corpus (Magnini et al. 2020, 2021) includes clinical cases in five different languages: Italian, English, French, Spanish, and Basque. Clinical cases are narratives written at the time of the medical visit that contain the symptoms, signs, diagnosis, treatment, and follow-up of an individual patient.
The clinical narratives were collected either from publications, like PubMed (journal abstracts) and The Pan African Medical Journal (journal articles), or from existing corpora like the SPACCC corpus (dataset). Other documents were collected from admission tests for specialties in medicine and abstracts of theses in medical science. The procedure used to collect the clinical narratives was conducted in different ways depending on the type of document and resource. For example, some of the documents in the English and French data were automatically extracted from PubMed through the PubMed API. In order to restrict the query to abstracts of clinical cases, the article category “clinical case” was selected in the API call. The documents extracted with this procedure were checked by human annotators to verify that the contents of these documents correspond to the definition of clinical case given in E3C. Such documents were finally split into three different sets (called layers), each of them containing its own clinical cases without any intersection between the clinical cases of two different layers.
-
• Layer 1: consists of clinical case texts with over 25K tokens per language. These texts include manual annotations of clinical entities and temporal information. Clinical entities (i.e., disorders like diseases or syndromes, findings, injuries or poisoning and signs or symptoms) are annotated at mention and concept level (Figure 3). A limitation of the mention-level annotation in E3C is that it does not specify which words inside a discontinuous entity span are actually part of the entity or not. For example, the entity respiratory signs in the text respiratory, digestive, laryngeal, vascular, or neurologic signs has been annotated by tagging the whole text span. This limitation is largely due to the tool (WebAnnoFootnote a) used to perform the manual annotation and which does not allow the annotation of discontinuous entities. Regarding the concept-level annotation, the disorder entities are mapped to their concept unique identifiers (CUIs) in UMLS. If an entity was not found in UMLS, then it was labeled CUI-less. The average inter-annotator agreement for the mention- and concept-level annotations is 75.00 (
 $F_1$
measure) and 91.00 (accuracy), respectively. Temporal information and factuality are events, time expressions, and temporal relations according to the THYME standard (Styler IV et al. Reference Styler, Bethard, Finan, Palmer, Pradhan, de Groen, Erickson, Miller, Lin, Savova and Pustejovsky2014). Table 1 presents a comprehensive overview of the data. It includes the exact counts of documents, sentences, and tokens per language, as well as the number of documents per language and source type. Additionally, the table highlights that only very few entities are discontinuous or nested.
$F_1$
measure) and 91.00 (accuracy), respectively. Temporal information and factuality are events, time expressions, and temporal relations according to the THYME standard (Styler IV et al. Reference Styler, Bethard, Finan, Palmer, Pradhan, de Groen, Erickson, Miller, Lin, Savova and Pustejovsky2014). Table 1 presents a comprehensive overview of the data. It includes the exact counts of documents, sentences, and tokens per language, as well as the number of documents per language and source type. Additionally, the table highlights that only very few entities are discontinuous or nested. -
• Layer 2: over 50K tokens of clinical case texts automatically annotated for clinical entities. The annotated entities were produced with a dictionary lookup method that matches the clinical entities in the text with the disorders in UMLS. A manual assessment of the quality of these annotated entities would be too demanding in terms of human resources. For this reason, the quality of Layer 2 was estimated through an indirect evaluation using the results obtained by the dictionary lookup method on Layer 1 (see Table 7).
-
• Layer 3: over 1 M tokens of clinical case texts or other medical texts with no annotations to be exploited by semi-supervised approaches.
Table 1. Layer 1: document, sentence, and token counts; source type distribution; entities by type and language.
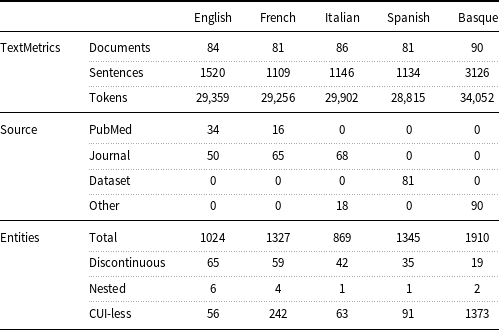
Table 2. Number of documents and disorder entities in the training and test partitions of Layer 1.
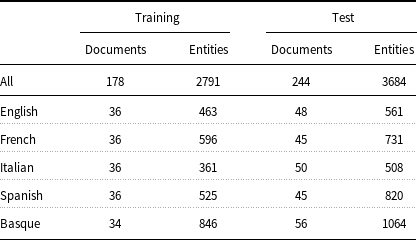

Figure 3. Example of annotated clinical case text. Each clinical entity is annotated with its name span and associated with its corresponding CUI in UMLS. The annotation does not specify which words inside a discontinuous entity span are actually part of the entity or not.
To let researchers compare their models under the same experimental conditions, Layer 1 has two partitions: one for training purposes (about 10K tokens) and one for testing (about 15K tokens) (Table 2). The reason for having a testing partition larger than that of training is that larger test datasets ensure a more accurate calculation of model performance. As regards Layer 2, researchers are free to use its automatically annotated entities in addition to the manual annotated entities in the training partition of Layer 1 for training their models.
4. Methods
One of the purposes of the present work is to establish the appropriateness of the clinical entity annotation of the E3C corpus to train an information extraction system for DNER. To do this, the training and test partitions of Layer 1 of the E3C corpus v2.0.0Footnote
b
have been used. A CRF model is set as the baseline to compare state-of-the-art pre-trained models to a traditional ML model. All the evaluated models were configured splitting the training partition randomly into two parts: a portion (80% of the documents) for training and a portion (20%) for tuning the models (development set). The resulting best configuration of each model was tested on the test partition. To assess the performance of the models, a standard
![]() $F_1$
-score has been used, which is a combination of precision and recall (Rijsbergen Reference Rijsbergen1979). The experiments were performed with Google Colab,Footnote
c
a free cloud-based service that allows the execution of Python code. One limitation of using Google Colab is that users who have recently used more resources in Colab are likely to run into usage limits and have their access to GPUs temporarily restricted.
$F_1$
-score has been used, which is a combination of precision and recall (Rijsbergen Reference Rijsbergen1979). The experiments were performed with Google Colab,Footnote
c
a free cloud-based service that allows the execution of Python code. One limitation of using Google Colab is that users who have recently used more resources in Colab are likely to run into usage limits and have their access to GPUs temporarily restricted.
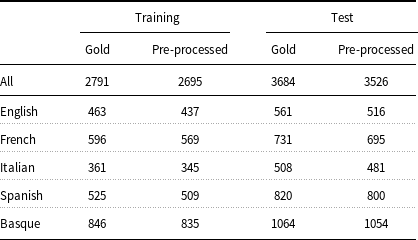
Table 3. Number of annotated entities before and after data preprocessing.
4.1. Preprocessing
Training and test data must be converted to an appropriate format before feeding into ML models. Typically, models for entity recognition require input data to be in IOB format and the models in the present work are no exception. In turn, to generate the IOB format, the input data must be tokenized and split into sentences. Even though the E3C corpus has already been pre-tokenized and sentence segmented, its documents are distributed in a format (UIMA CAS XMI) that has to be transformed into IOB before being used by the models. Unfortunately, the IOB format cannot be adopted to represent discontinuous or nested entities, which are both present in the corpus. Concerning the representation of the discontinuous entities (3.4% of the total entities in the corpus), some extensions to the IOB scheme have been proposed, such as the scheme of Tang et al. (Reference Tang, Wu, Jiang, Denny and Xu2013). This scheme requires the tokens inside a discontinuous entity to be known exactly. Since the mention-level annotation of the corpus does not provide such information for the discontinuous entities (see Section 3), this kind of entities has been removed from consideration. As far as the nested entities are concerned, it has been observed that there are few of them in the corpus (0.2% of the entities). For this reason, only the topmost entities have been considered. Another issue that had to be addressed was related to the character encoding of a document (IT101195). Given that it was not possible to parse this document correctly, the latter has been discarded from the corpus. Table 3 shows that, despite these shortcomings, 96.6% (2695 out of 2791) of the entities in the training partition and 95.7% (3526 out of 3684) of those in the test partition have been preserved after data preprocessing. The pre-processed data have been made available through the GitLab repository.Footnote d
4.2. Evaluation of the models
CRF (Lafferty et al. Reference Lafferty, McCallum and Pereira2001) is a well-known ML method that has been widely used in NER (McCallum and Li Reference McCallum and Li2003). In the present work, the CRF method has been implemented via an adaptation of the code by Korobov (Reference Korobov2021). For each running word, the (lowercase) word itself and prefixes and suffixes (1, 2, 3, 4 characters at the start/end of the word) have been used as features. Each of these features has been extracted for the current, previous, and following (lowercase) words. With this procedure, the CRF modelFootnote e has been trained and evaluated on each language of the corpus.
Traditional ML models like CRF usually require a large amount of data to achieve high performance. Unfortunately, available annotated datasets for DNER, including the E3C corpus, only consist of a few hundred thousand annotated words.
Transfer learning is a ML technique that helps users overcome scarcity of labeled data by reusing models pre-trained on large datasets as the starting point to build a model for a new target task. In his 2021 book, Azunre (Reference Azunre2021) expressed this concept as follows: “Transfer Learning enables you to adapt or transfer the knowledge acquired from one set of tasks and/or domains, to a different set of tasks and/or domains. What this means is that a model trained with massive resources—including data, computing power, time, cost, etc.—once open-sourced can be fine-tuned and re-used in new settings by the wider engineering community at a fraction of the original resource requirements.”
In an attempt to exploit the ability of pre-trained models like RoBERTa to achieve better results than other ML models on small datasets, the RoBERTa and BERT models have been compared. As these models are pre-trained on English data, they have been fine-tuned and evaluated on the English portion of the corpus only.
In the present study, the XLM-RoBERTa model has been tested to take advantage of the multilingual annotation of the corpus. This is possible because multilingual models such as XLM-RoBERTa are pre-trained on the corpus from multiple languages and hence they can be used for NER tasks in more than one language. XLM-RoBERTa has been evaluated in three different settings. In the first setting, the model has been tested on each target language data by fine-tuning on data in the target language. This evaluates per-language performance. In the second setting, the models have been fine-tuned on data in all languages to evaluate multilingual learning. In the third setting, the models have been fine-tuned on data in all languages except the target language, and performance has been evaluated on the target language. In this way, the exploitability of the E3C corpus has been evaluated for cross-lingual transfer learning.
Since the XLM-RoBERTa model comes pre-trained on generic corpora, its performance may be limited when the model is used to annotate clinical case texts. To see how much this model compared to state-of-the-art models in the biomedical domain, the English portion of the E3C corpus has been used to compare XLM-RoBERTa with the BERN2, HunFlair, and BioBERT models.
Unlike Layer 1, which contains manual annotated entities, Layer 2 consists of automatically annotated entities. For the purpose of investigating the appropriateness of Layer 2 for training ML models, documents from Layer 1 and Layer 2 have been concatenated into one larger train dataset. Then, the XLM-RoBERTa model was fine-tuned on such a dataset.
The setup used to evaluate all the pre-trained models mentioned above (except HunFlair for which its own scripts were usedFootnote f ) is essentially the same as that implemented in dl blog (2021), which uses the Python script run_ner.pyFootnote g to execute its code.
One of the main hyper-parameters that may affect the accuracy of pre-trained models is the number of learning epochs. In fact, while too many epochs can lead to overfitting of the training dataset, too few epochs may result in an underfitting model. The optimal number of epochs for fine-tuning pre-trained models is generally low. For example, the authors of BERT recommend only 2–4 epochs. Taking these considerations into account, the loss function calculated on the development partition of the corpus has been used to detect when a model is overfitting. A good fit is when the loss function stops improving after a certain number of epochs and begins to decrease afterward. In this study, the search of the optimal number of epochs was limited between 2 and 4 to avoid going beyond the application usage limits of Google Colab. For most of the evaluated models, the optimal number of epochs was found equal to 4. For the rest of the models, the observed optimal number of epochs was 3 without, however, a significant difference in the loss gap between 3 and 4 epochs. For this reason, all the models have been fine-tuned with 4 epochs as shown in Table 4.
Table 4. Deep learning models and hyper-parameters used in the setup.
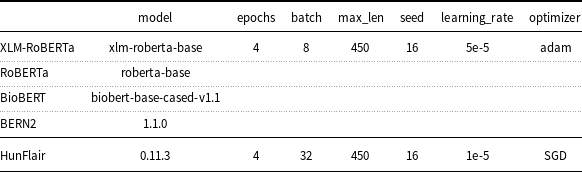
Transformers v3.1.0 was used to perform the experiments except for BERN2, which was evaluated with transformers v4.9.0.
To ensure the reproducibility of results, the random seed hyper-parameter was set to a fixed value (16). Since the choice of the seed value can result in substantial differences in scores (Reimers and Gurevych Reference Reimers and Gurevych2017), each experiment was repeated 30 times, varying the seed each time. This set of experiments was conducted with a dedicated PCFootnote h to overcome the computational time limitation of Google Colab. Eventually, statistical differences between two models were calculated using the paired permutation test (Noreen Reference Noreen1989; Dror et al. Reference Dror, Baumer, Shlomov and Reichart2018). This was done by using the permutation_test functionFootnote i of the MlxtendFootnote j python library.
The results of the considered models have been compared with those of two baseline methods often used in the literature. A first baseline was produced by a system that only identifies entities appearing in the training split of the corpus. This baseline was used at CoNLL-2003 in the task of NER. The second baseline finds an exact string match for each disorder name in UMLS to a word or phrase in each document of the test. This baseline owes a lot to the one used by Jonnagaddala et al. (Reference Jonnagaddala, Jue, Chang and Dai2016). The only difference is that in this work UMLS has been used as a controlled vocabulary instead of the MEDIC vocabulary (Davis et al. Reference Davis, Wiegers, Rosenstein and Mattingly2012).
The reason why state-of-the-art NLP tools like cTAKES and MetaMap have not been included among these baselines is that these tools are often only available for English and they are not easily adaptable to other languages such as the ones of the E3C corpus.
To provide researchers with benchmarking baselines on the concept-level annotation of the corpus, entity linking has been implemented in cascade to the best performing model (XLM-RoBERTa). The approach for entity linking used here is practically the same as the one proposed by Alam et al. (Reference Alam, Corazza, Lavelli and Zanoli2016). Specifically, in the present work a dictionary lookup based on UMLS has been adopted instead of the Comparative Toxicogenomics Database to select the best-matching concept (see the pseudocode in Algorithm 1). UMLS consists of more than 100 source vocabularies, including SNOMED CT, which is subject to license restriction when it is used in SNOMED nonmember countries like Italy. This restriction has been addressed by removing SNOMED CT from the experimentation. The results using the presented approach for entity linking have been compared with the results of the two baselines that have also been used to evaluate DNER: the CoNLL-2003 and dictionary lookup baselines. In particular, in order to evaluate entity linking, the baselines have been configured in such a way that they only select complete unambiguous entities appearing in the training data (CoNLL-2003) or in UMLS (dictionary lookup). The standard metric used to evaluate the linked entities is the metric used for NER (
![]() $F_1$
-score). Particularly, in entity linking tasks an entity link is considered correct only if the entity matches the gold boundary and the link to the entity is also correct.
$F_1$
-score). Particularly, in entity linking tasks an entity link is considered correct only if the entity matches the gold boundary and the link to the entity is also correct.
Table 5.
![]() $F_1$
measure of the machine learning models and baselines (dictionary lookup, CoNLL-2003) on the mention-level test set.
$F_1$
measure of the machine learning models and baselines (dictionary lookup, CoNLL-2003) on the mention-level test set.
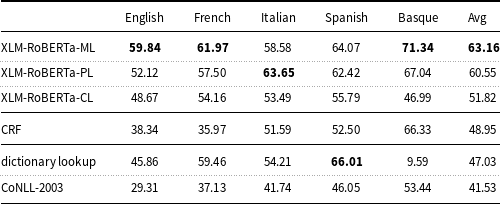
XLM-RoBERTa is fine-tuned and evaluated on all languages (-ML), per-language (-PL), and cross-language (-CL). The results of the deep learning models were computed with a fixed random seed (16).
Algorithm 1. Pseudocode for concept normalization. pred_entities are the entities recognized by the XLM-RoBERTa model. gold_entities, UMLS are dictionaries in which disorder entities are associated with their concept unique identifiers (CUIs) in UMLS.

5. Results
Table 5 shows that all the pre-trained models, which are based on XLM-RoBERTa, outperform the CoNLL-2003 baseline and perform better on average than the dictionary lookup baseline. The results of these models are also higher than the results of the traditional ML model (CRF).
The XLM-RoBERTa-ML model fine-tuned on all language data simultaneously (multilingual learning) performs better (except for Italian) than XLM-RoBERTa-PL fine-tuned on each language data separately (per-language learning), with a
![]() $F_1$
measure of 63.16 and 60.55, respectively.
$F_1$
measure of 63.16 and 60.55, respectively.
XLM-RoBERTa-CL fine-tuned on all language data except the target language and evaluated on the target language (cross-lingual transfer learning) produces lower results for Basque (
![]() $F_1$
measure: 46.99) than for the other languages of the corpus. Finally, UMLS, used to implement the dictionary lookup baseline, has a low coverage in Basque (
$F_1$
measure: 46.99) than for the other languages of the corpus. Finally, UMLS, used to implement the dictionary lookup baseline, has a low coverage in Basque (
![]() $F_1$
measure: 9.59).
$F_1$
measure: 9.59).
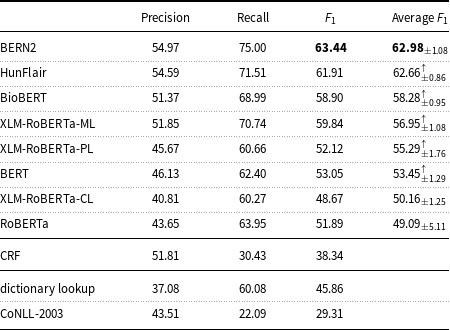
On the English data, Table 6 highlights that the results of the state-of-the-art BNER tools such as BERN2 and HunFlair are substantially better than the results of the other tested models. The BERN2 result (
![]() $F_1$
measure: 63.44) is higher than that of the median for all systems (
$F_1$
measure: 63.44) is higher than that of the median for all systems (
![]() $F_1$
measure: 58.90) participating in Task 1 of the ShARe/CLEF eHealth Evaluation Lab 2013, but lower than the best result in Task 1 (
$F_1$
measure: 58.90) participating in Task 1 of the ShARe/CLEF eHealth Evaluation Lab 2013, but lower than the best result in Task 1 (
![]() $F_1$
measure: 75.00).
$F_1$
measure: 75.00).
Table 6. Precision, recall, and
![]() $F_1$
measure of the machine learning models and baselines (dictionary lookup, CoNLL-2003) on the English mention-level test set.
$F_1$
measure of the machine learning models and baselines (dictionary lookup, CoNLL-2003) on the English mention-level test set.
Average
![]() $F_1$
measure and standard deviation are calculated over 30 random seeds [1–30]. (
$F_1$
measure and standard deviation are calculated over 30 random seeds [1–30]. (
![]() $\uparrow$
) indicates that the average
$\uparrow$
) indicates that the average
![]() $F_1$
measure of the model is significantly better than the one immediately below for p-value
$F_1$
measure of the model is significantly better than the one immediately below for p-value
![]() $\leq$
0.05.
$\leq$
0.05.
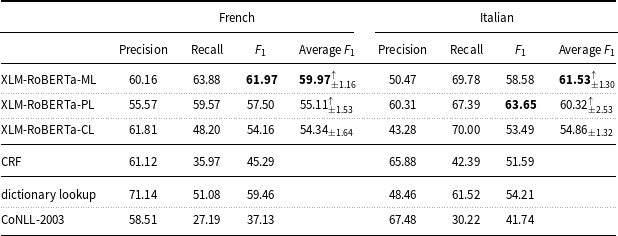
With regard to the tradeoff between precision and recall, all models based on deep learning show higher recall values than precision values on the English and Italian data (Tables 6 and 7), while they show a closer balance between precision and recall on the French, Spanish, and Basque data (Tables 7 and 8).
Table 7. Precision, recall, and
![]() $F_1$
measure of the machine learning models and baselines (dictionary lookup, CoNLL-2003) on the French and Italian mention-level test sets.
$F_1$
measure of the machine learning models and baselines (dictionary lookup, CoNLL-2003) on the French and Italian mention-level test sets.
Average
![]() $F_1$
measure and standard deviation are calculated over 30 random seeds [1–30]. (
$F_1$
measure and standard deviation are calculated over 30 random seeds [1–30]. (
![]() $\uparrow$
) indicates that the average
$\uparrow$
) indicates that the average
![]() $F_1$
measure of the model is significantly better than the one immediately below for p-value
$F_1$
measure of the model is significantly better than the one immediately below for p-value
![]() $\leq$
0.05.
$\leq$
0.05.
Table 8. Precision, recall, and
![]() $F_1$
measure of the machine learning models and baselines (dictionary lookup, CoNLL-2003) on the Spanish and Basque mention-level test sets.
$F_1$
measure of the machine learning models and baselines (dictionary lookup, CoNLL-2003) on the Spanish and Basque mention-level test sets.

Average
![]() $F_1$
measure and standard deviation are calculated over 30 random seeds [1–30]. (
$F_1$
measure and standard deviation are calculated over 30 random seeds [1–30]. (
![]() $\uparrow$
) indicates that the average
$\uparrow$
) indicates that the average
![]() $F_1$
measure of the model is significantly better than the one immediately below for p-value
$F_1$
measure of the model is significantly better than the one immediately below for p-value
![]() $\leq$
0.05.
$\leq$
0.05.
On the French data (Table 7), XLM-RoBERTa-PL (
![]() $F_1$
measure: 57.50) and CRF (
$F_1$
measure: 57.50) and CRF (
![]() $F_1$
measure: 45.29) perform in line with multilingual BERT and CRF tested by the participants of Task 3 at DEFT 2020 (
$F_1$
measure: 45.29) perform in line with multilingual BERT and CRF tested by the participants of Task 3 at DEFT 2020 (
![]() $F_1$
measure: 53.03 and 49.84, respectively).
$F_1$
measure: 53.03 and 49.84, respectively).
As far as the exploitation of Layer 2 for training the models is concerned, fine-tuning XLM-RoBERTa on the concatenation of documents of Layer 1 and Layer 2 produces lower results than when XLM-RoBERTa is fine-tuned on Layer 1 only (Table 9).
Table 9. Precision, recall, and
![]() $F_1$
measure of XLM-RoBERTa-ML fine-tuned on the concatenation of documents of Layer 1 and Layer 2 compared to XLM-RoBERTa-ML fine-tuned using documents of Layer 1.
$F_1$
measure of XLM-RoBERTa-ML fine-tuned on the concatenation of documents of Layer 1 and Layer 2 compared to XLM-RoBERTa-ML fine-tuned using documents of Layer 1.
Concerning entity linking, Table 10 shows that the proposed approach (
![]() $F_1$
measure: 50.37) performs much better than the dictionary lookup (40.93) and CoNLL-2003 (39.0) baselines. It also performs comparable on average with the results of the dictionary lookup baseline method used at the BC5CDR task (
$F_1$
measure: 50.37) performs much better than the dictionary lookup (40.93) and CoNLL-2003 (39.0) baselines. It also performs comparable on average with the results of the dictionary lookup baseline method used at the BC5CDR task (
![]() $F_1$
measure: 52.30) and better than the dictionary lookup baseline on the NCBI dataset (
$F_1$
measure: 52.30) and better than the dictionary lookup baseline on the NCBI dataset (
![]() $F_1$
measure: 33.10).
$F_1$
measure: 33.10).
Table 10.
![]() $F_1$
measure of entity linking in cascade to XLM-RoBERTa-ML (the approach of the present work) and baselines (dictionary lookup and CoNLL-2003) calculated on the concept-level annotation of the corpus. For each column, the highest value of among the three approaches is displayed in bold.
$F_1$
measure of entity linking in cascade to XLM-RoBERTa-ML (the approach of the present work) and baselines (dictionary lookup and CoNLL-2003) calculated on the concept-level annotation of the corpus. For each column, the highest value of among the three approaches is displayed in bold.

In brackets the accuracy measure, which is calculated as the number of correctly linked entities divided by the total number of all correctly recognized entities.
One of the main outcomes of this study is the integration of the considered models into the ELG platform, a cloud platform providing easy access to hundreds of commercial and noncommercial language technology resources (tools, services, and datasets) for all European languages. To do this, a pipeline has been implemented, which consists of the best model for DNER (XLM-RoBERTa-ML) in combination with the described approach for entity linking. Then, the pipeline has been packed as a docker image and deployed on the platform as an ELG service.Footnote k This allows users to experiment with the pipeline in three different ways: (i) trying the pipeline from its web page in ELG (Figure 4), (ii) running the pipeline by command line from their shell (Figure 5), and (iii) using the pipeline from their Python code by exploiting the ELG Python SDK. The pre-processed datasets used for training and testing the models are hosted on GitLab.Footnote l
Figure 4. Running the pipeline for DNER from its page in the ELG platform.

Figure 5. Running the pipeline for DNER from the user’s shell.
6. Discussion
Machines are much faster at processing knowledge compared to humans, but they require manually annotated datasets for training. The overall outcome of the experiments shows that the E3C corpus can be used to successfully train and evaluate ML models for DNER.
One of the main problems that had to be faced was related to the preprocessing of the E3C corpus. In fact, the corpus contains both discontinuous and nested entities that cannot be addressed using the classical IOB tagging scheme (Section 4.1). Although there are extensions to the IOB schema capable of encoding discontinuous entities, they cannot be applied to the E3C corpus due to the lack of required information in the annotation of the corpus. For this reason, the discontinuous entities have been removed from consideration (3.4% of the total number of entities in the corpus). Concerning the nested entities, given the relatively small number of them in the corpus (0.2%) and the difficulty of working with formats other than IOB, it was decided to use the topmost entities of the corpus only.
Regarding the effectiveness of the considered models, Table 5 compares the results of the ML models trained on the mention-level annotation with the results of two baselines often used in the literature. Significantly, the ML models outperform the CoNLL-2003 baseline, since the latter only identified entities seen in the training data. This suggests that the E3C corpus can be used to train models that generalize well on unseen data.
Surprisingly, higher values for multilingual learning (XLM-RoBERTa-ML) than for training on each language separately (XLM-RoBERTa-PL) have been detected (with a
![]() $F_1$
measure of 63.16 and 60.55, respectively) (Table 5), whereas Conneau et al. (Reference Conneau, Khandelwal, Goyal, Chaudhary, Wenzek, Guzmán, Grave, Ott, Zettlemoyer and Stoyanov2020) found no substantial differences between the two learning approaches on the CoNLL datasets (with a
$F_1$
measure of 63.16 and 60.55, respectively) (Table 5), whereas Conneau et al. (Reference Conneau, Khandelwal, Goyal, Chaudhary, Wenzek, Guzmán, Grave, Ott, Zettlemoyer and Stoyanov2020) found no substantial differences between the two learning approaches on the CoNLL datasets (with a
![]() $F_1$
measure of 89.43 and 90.24, respectively). This points toward the idea that the five languages data of the E3C corpus can be put together to form a larger partition and this improves the accuracy of the trained models. The reasons for contradictory results with the Italian data depend on the specific random seed value used for experimentation as discussed later in this section.
$F_1$
measure of 89.43 and 90.24, respectively). This points toward the idea that the five languages data of the E3C corpus can be put together to form a larger partition and this improves the accuracy of the trained models. The reasons for contradictory results with the Italian data depend on the specific random seed value used for experimentation as discussed later in this section.
The experiments conducted to evaluate cross-lingual transfer (XLM-RoBERTa-CL) (Table 5) validate the appropriateness of the corpus to train models on data available for one language to recognize disorder entities in another language. As expected, the highest accuracy values were obtained with those models fine-tuned and tested on the Romance languages of the corpus (Italian, Spanish, and French), all of which stem from Latin. On the other hand, Basque has no Latin base. Consequently, it is not surprising that training on Romance languages and testing on Basque did not produce optimal results. Despite these not ideal results for Basque, however, they are considerably higher than the results produced by the dictionary lookup baseline. This shows that the E3C corpus is also applicable to low-resource languages that have no training data and with a low level of coverage in medical vocabularies.
Turning now to the comparison between deep learning models and more traditional ML models, much higher values for the pre-trained models (XLM-RoBERTa) than CRF (Table 5) have been found. This fact is explainable by considering that pre-trained models allow for fine-tuning this task on a much smaller dataset than would be required in a model that is built from scratch like CRF.
Table 6 indicates that state-of-the-art tools for BNER like BERN2 and HunFlair outperform the other tested models.
With regard to how the considered models compare to models evaluated on other datasets for BNER, the results obtained with BioBERT (
![]() $F_1$
measure: 58.90) on the English data of the corpus are lower than the results achieved by the BioBERT authors on the NCBI (
$F_1$
measure: 58.90) on the English data of the corpus are lower than the results achieved by the BioBERT authors on the NCBI (
![]() $F_1$
measure: 89.71) and BC5CDR (
$F_1$
measure: 89.71) and BC5CDR (
![]() $F_1$
measure: 87.15) datasets. Then, the results on the English data are also lower than the results that the authors of this work achieved by evaluating BioBERT on the dataset used in Task 1 of the ShARe/CLEF eHealth Evaluation Lab 2013 (
$F_1$
measure: 87.15) datasets. Then, the results on the English data are also lower than the results that the authors of this work achieved by evaluating BioBERT on the dataset used in Task 1 of the ShARe/CLEF eHealth Evaluation Lab 2013 (
![]() $F_1$
measure: 82.02). This non-ideal performance on the E3C corpus was not completely unexpected. In fact, the experiments carried out also show lower results of the CoNLL-2003 baseline on the E3C English data (
$F_1$
measure: 82.02). This non-ideal performance on the E3C corpus was not completely unexpected. In fact, the experiments carried out also show lower results of the CoNLL-2003 baseline on the E3C English data (
![]() $F_1$
measure: 29.31) than of those of the CoNLL-2003 baseline on the NCBI (
$F_1$
measure: 29.31) than of those of the CoNLL-2003 baseline on the NCBI (
![]() $F_1$
measure: 69.01) and BC5CDR (
$F_1$
measure: 69.01) and BC5CDR (
![]() $F_1$
measure: 69.22) datasets. On the E3C English data, the CoNLL-2003 baseline also produces considerably lower results than the CoNLL-2003 baseline tested on the ShARe/CLEF dataset (
$F_1$
measure: 69.22) datasets. On the E3C English data, the CoNLL-2003 baseline also produces considerably lower results than the CoNLL-2003 baseline tested on the ShARe/CLEF dataset (
![]() $F_1$
measure: 51.03). This implies that many entities are shared between the training and test partitions of the compared datasets and suggest why the models tested on the E3C corpus perform far differently than the models tested on the other datasets. Then, it is interesting to note that the ML models trained on the E3C corpus achieved one of the highest classification accuracy values in comparison with the CoNLL-2003 baseline. This result would seem to support the hypothesis that the patterns learned from E3C can help recognize new entities not seen during the training of models.
$F_1$
measure: 51.03). This implies that many entities are shared between the training and test partitions of the compared datasets and suggest why the models tested on the E3C corpus perform far differently than the models tested on the other datasets. Then, it is interesting to note that the ML models trained on the E3C corpus achieved one of the highest classification accuracy values in comparison with the CoNLL-2003 baseline. This result would seem to support the hypothesis that the patterns learned from E3C can help recognize new entities not seen during the training of models.
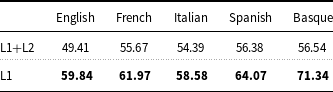
As far as the results obtained on the French data are concerned (Table 7), XLM-RoBERTa-ML (
![]() $F_1$
measure: 61.97) performed in line with the second best system (
$F_1$
measure: 61.97) performed in line with the second best system (
![]() $F_1$
measure: 61.41) in Task 3 of the DEFT 2020 challenge (Cardon et al. Reference Cardon, Grabar, Grouin and Hamon2020) on the sub-task of identifying pathologies and signs or symptoms (disorders in E3C). This system was based on a hybrid architecture of LSTM + CRF in cascade to BERT. Then, XLM-RoBERTa-PL (
$F_1$
measure: 61.41) in Task 3 of the DEFT 2020 challenge (Cardon et al. Reference Cardon, Grabar, Grouin and Hamon2020) on the sub-task of identifying pathologies and signs or symptoms (disorders in E3C). This system was based on a hybrid architecture of LSTM + CRF in cascade to BERT. Then, XLM-RoBERTa-PL (
![]() $F_1$
measure: 57.50) and CRF (
$F_1$
measure: 57.50) and CRF (
![]() $F_1$
measure: 45.29) perform in line with multilingual BERT (
$F_1$
measure: 45.29) perform in line with multilingual BERT (
![]() $F_1$
measure: 53.03) and CRF (
$F_1$
measure: 53.03) and CRF (
![]() $F_1$
measure: 49.84) tested by the participant teams in Task 3 (Copara et al. Reference Copara, Knafou, Naderi, Moro, Ruch and Teodoro2020). It is important to note that the performance of the models on the DEFT dataset is lower than that of the models on the other datasets discussed before, but also that the CoNLL-2003 baseline (
$F_1$
measure: 49.84) tested by the participant teams in Task 3 (Copara et al. Reference Copara, Knafou, Naderi, Moro, Ruch and Teodoro2020). It is important to note that the performance of the models on the DEFT dataset is lower than that of the models on the other datasets discussed before, but also that the CoNLL-2003 baseline (
![]() $F_1$
measure: 16.35) on the DEFT dataset is lower than the CoNLL-2003 baseline on such datasets.
$F_1$
measure: 16.35) on the DEFT dataset is lower than the CoNLL-2003 baseline on such datasets.
When computational resources are limited, random seed is one of those hyper-parameters that are often kept constant to reduce the number of system configurations to evaluate. To see how the random seed setup can affect the model performance, the results of the pre-trained models calculated with one fixed random seed were compared to the results of the models calculated over 30 different random seeds. Significantly, the average
![]() $F_1$
measure computed over the 30 random seeds (Tables 6
–8) confirms the results obtained with a fixed random seed that the ML models outperform the considered baselines. The observations made on how the considered models compare to models evaluated on other datasets are also confirmed. Looking at how the models compare to each other, XLM-RoBERTa-ML was thought initially better than BioBERT on the English data, while XLM-RoBERTa-PL was considered better than XLM-RoBERTa-ML on the Italian data. However, a more careful analysis based on the average
$F_1$
measure computed over the 30 random seeds (Tables 6
–8) confirms the results obtained with a fixed random seed that the ML models outperform the considered baselines. The observations made on how the considered models compare to models evaluated on other datasets are also confirmed. Looking at how the models compare to each other, XLM-RoBERTa-ML was thought initially better than BioBERT on the English data, while XLM-RoBERTa-PL was considered better than XLM-RoBERTa-ML on the Italian data. However, a more careful analysis based on the average
![]() $F_1$
measure revealed that these results were due to the fixed random seed used in the experimentation. This stress the importance of testing deep learning models with many random seeds, but also that this often requires expensive hardware and extensive computational costs.
$F_1$
measure revealed that these results were due to the fixed random seed used in the experimentation. This stress the importance of testing deep learning models with many random seeds, but also that this often requires expensive hardware and extensive computational costs.
The exploitability of Layer 2 for training ML models is discussed as follows. The study carried out was not successful in proving its usefulness for improving NER (Table 9). However, this may depend on the methodology chosen for this experiment. In fact, documents from Layer 1 and Layer 2 have been concatenated into one large dataset for training the models. However, the distant supervision method used to annotate Layer 2 may have induced incomplete and noisy labels, making the straightforward application of supervised learning ineffective. It is the authors’ opinion that using heuristic rules to filter out sentences with potentially low matching quality (e.g. sentences that contain other entities besides the entities annotated in the training partition of Layer 1) might be beneficial for using Layer 2 successfully.
In an attempt to provide researchers with baselines on the concept-level annotation of the corpus, a dictionary lookup method has been implemented in cascade to the model for DNER that on average performed better than others on all five languages of the corpus (XLM-RoBERTa-ML). This approach performs largely in line with the dictionary lookup baseline method but better than the CoNLL-2003 baseline (Table 9). It also performs comparably on average with the dictionary lookup baseline used at the BC5CDR task (
![]() $F_1$
measure: 52.30) and better than the dictionary lookup baseline on the NCBI dataset (
$F_1$
measure: 52.30) and better than the dictionary lookup baseline on the NCBI dataset (
![]() $F_1$
measure: 33.10).
$F_1$
measure: 33.10).
The authors of the present work are confident that their findings may be useful for people to better understand the appropriateness of the E3C corpus for training DNER models. The authors believe that the trained models might also be applied to unseen languages that are not covered by any language of the corpus but that share grammatical structures and patterns with them. These models might also represent a valuable solution for low-resource languages for which does not exist any annotated data. Researchers can use these results as the baselines for this corpus to develop and compare their own models. The distribution of the considered models through the ELG platform enables clinical researchers and practitioners to have quick and easy access to these models.
7. Conclusion
This work has discussed experiments carried out on the E3C corpus of biomedical annotations, showing the appropriateness of the corpus itself for training ML models and developing a system capable of mining entities of disorders from clinical case texts. The results achieved in this regard form a first baseline that researchers can use to compare their results and systems with. Clinical researchers and practitioners can experiment with the resulting models via the ELG platform.
Acknowledgments
This work has been partially funded by the European Language Grid (ELG) project (EU grant no. 825627). We acknowledge the support of the PNRR project FAIR - Future AI Research (PE00000013), under the NRRP MUR program funded by the NextGenerationEU. The authors would like to thank the ELG team for their support with the tool integration in the ELG platform. A special thank goes to Fabio Rinaldi and Begoña Altuna for their comments and valuable suggestions. Thanks are also due to Dr. Natalia Grabar and Dr. Guergana Savova for helping us obtain the datasets used at DEFT 2020 and ShARe/CLEF eHealth 2013, respectively.
Competing interests
The authors declare none.

























