Introduction
Lichen symbiosis is undergoing renewed scrutiny, in part because of the application of molecular ecology techniques such as metabarcoding (Hawksworth & Grube, Reference Hawksworth and Grube2020; Smith et al., Reference Smith, Dal Grande, Muggia, Keuler, Divakar, Grewe, Schmitt, Lumbsch and Leavitt2020). Historic assumptions about the ‘single fungus and single eukaryote alga or Cyanobacteria’ nature of the symbiosis do not take into account tripartite lichens (i.e., those containing cyanobacterial and algal photobionts) (Rikkinen et al., Reference Rikkinen, Oksanen and Lohtander2002; Henskens et al., Reference Henskens, Green and Wilkins2012), and have been further challenged by the more recent discovery of other fungi (Millanes et al., Reference Millanes, Diederich and Wedin2016; Spribille et al., Reference Spribille, Tuovinen, Resl, Vanderpool, Wolinski, Aime, Schneider, Stabentheiner, Toome-Heller, Thor, Mayrhofer, Johannesson and McCutcheon2016; Mark et al., Reference Mark, Laanisto, Bueno, Niinemets, Keller and Scheidegger2020), eukaryote algae (chlorobionts) and Cyanobacteria (cyanobionts) (Henskens et al., Reference Henskens, Green and Wilkins2012; Moya et al., Reference Moya, Molins, Martínez-Alberola, Muggia and Barreno2017) within lichens in addition to the primary myco- and photobionts (Smith et al., Reference Smith, Dal Grande, Muggia, Keuler, Divakar, Grewe, Schmitt, Lumbsch and Leavitt2020), alongside a complex diversity of heterotrophic bacteria (Grube et al., Reference Grube, Cardinale, de Castro, Müller and Berg2009). This increased diversity of organisms co-existing within the lichen thallus has added support to an ongoing reinterpretation of lichens as even more complex ecological associations comprising numerous interacting components (Honegger, Reference Honegger1991; Hawksworth & Grube, Reference Hawksworth and Grube2020).
Amongst this complex lichen community, photobionts are fundamental members responsible for providing sugars (and in the case of some cyanobacterial photobionts, available nitrogen) essential for the survival of the lichen complex (holobiont). Extended photobiont diversity is likely widespread in terrestrial lichens (Dal Forno et al., Reference Dal Forno, Lawrey, Sikaroodi, Gillevet, Schuettpelz and Lücking2021) and has been hypothesized to be associated with increased ecological adaptive capability (Casano et al., Reference Casano, del Campo, García-Breijo, Reig-Armiñana, Gasulla, del Hoyo, Guéra and Barreno2011; del Campo et al., Reference del Campo, Catalá, Gimeno, del Hoyo, Martínez-Alberola, Casano, Grube and Barreno2013; Muggia et al., Reference Muggia, Vancurova, Škaloud, Peksa, Wedin and Grube2013; Ruprecht et al., Reference Ruprecht, Brunauer and Türk2014). By recruiting a diverse range of photobionts, it has been proposed that lichens may benefit from enhanced ecological plasticity, facilitating the ability to survive a range of environments (Casano et al., Reference Casano, del Campo, García-Breijo, Reig-Armiñana, Gasulla, del Hoyo, Guéra and Barreno2011; del Campo et al., Reference del Campo, Catalá, Gimeno, del Hoyo, Martínez-Alberola, Casano, Grube and Barreno2013; Muggia et al., Reference Muggia, Vancurova, Škaloud, Peksa, Wedin and Grube2013). For example, the soil crust lichen Psora decipiens supports multiple lineages of Trebouxia and Asterochloris allowing it to dominate soil crusts from deserts to alpine sites (Ruprecht et al., Reference Ruprecht, Brunauer and Türk2014). Additionally, photobiont ‘switching’ may allow for a further increase in fitness where multiple mycobiont lineages share locally adapted photobiont genotypes (Piercey-Normore & DePriest, Reference Piercey-Normore and DePriest2001).
Changing environmental conditions are characteristic of the habitat occupied by marine lichens in the intertidal zone, with marine lichens being exposed to a uniquely dynamic range of pressures not experienced by terrestrial lichens including daily tidal cycles of seawater coverage. Lichina pygmaea (Lightf.) C. Agardh is a saxicolous fruticose lichen in the family Lichinaceae that inhabits the upper intertidal zone (Figure 1A). It can be found throughout the North Atlantic coastline, including the area surrounding Plymouth, UK (Naylor, Reference Naylor1930). L. pygmaea was historically thought to form a symbiosis with Calothrix (Cyanobacteria) (Whitton, Reference Whitton2012), which was updated through molecular characterization to Rivularia (Ortiz-Álvarez et al., Reference Ortiz-Álvarez, de los Ríos, Fernández-Mendoza, Torralba-Burrial and Pérez-Ortega2015). Marine lichens such as L. pygmaea remain relatively under-studied compared with terrestrial taxa, and the degree to which complex photobiont diversity occurs in marine lichens is yet to be determined. The potential for multiple cyanobionts within members of the Lichinaceae was proposed by Bubrick & Galun (Reference Bubrick and Galun1984), but the extent of this has not yet been investigated using molecular ecology techniques. Here, we perform a broad metabarcoding survey of the bacterial (16S rRNA gene) and eukaryote (18S rRNA gene) phototrophic taxa associated with the L. pygmaea thallus to establish the potential for complex photobiont diversity in a marine lichen.
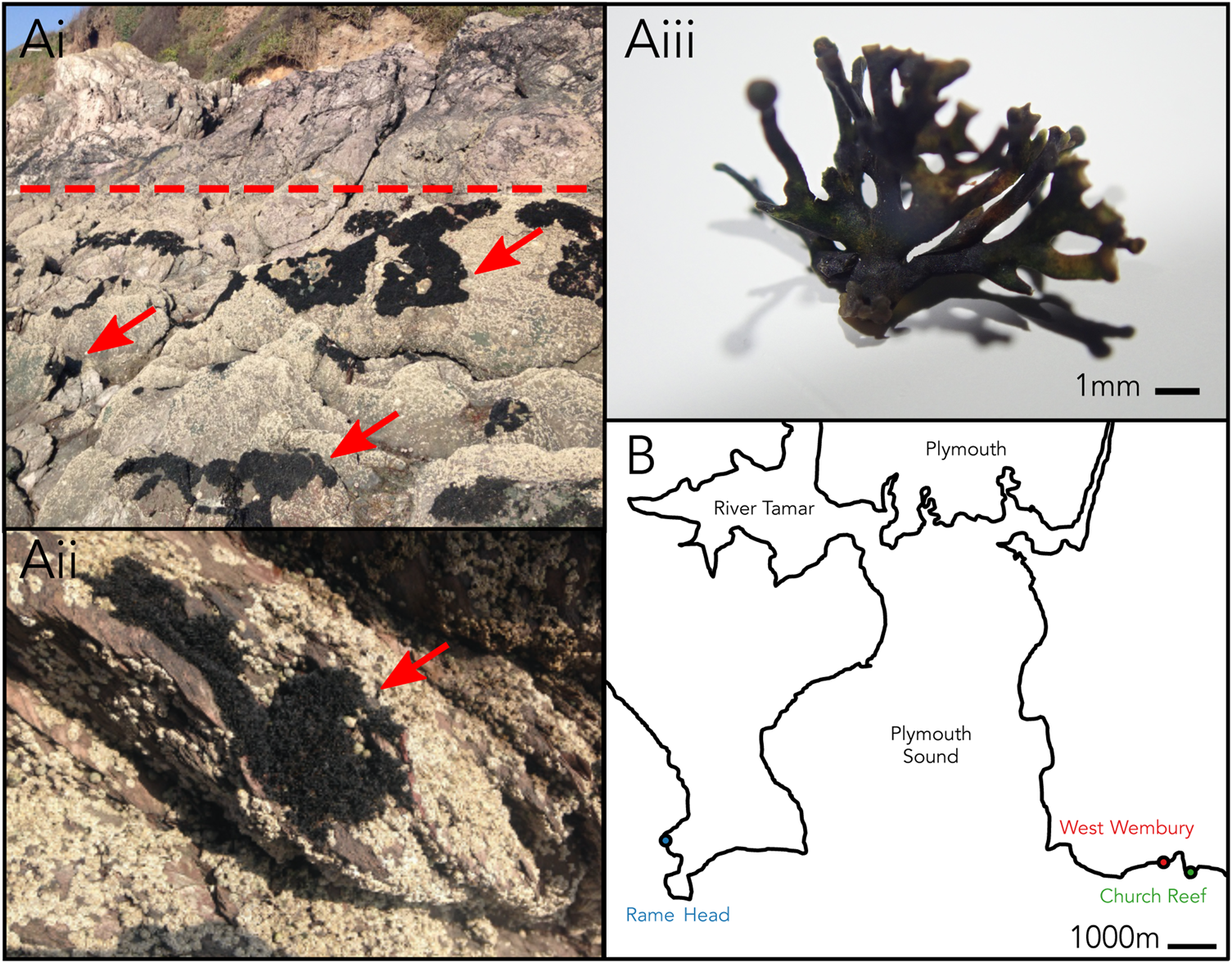
Fig. 1. (Ai–iii) Lichina pygmaea at Church Reef with close-up of thallus. Dotted line indicates approximate upper limit of the intertidal. Arrows point to L. pygmaea thalli. (B) Sampling locations of L. pygmaea near Plymouth (UK).
Methods
Sampling
Sampling and processing were carried out according to the protocol described in Parrot et al. (Reference Parrot, Antony-Babu, Intertaglia, Grube, Tomasi and Suzuki2015). In summary, L. pygmaea thalli were collected from three locations close to Plymouth in the south-west of the UK (Figure 1B) in spring 2016: Rame Head (lat = 50.3228, long = −4.2194), West Wembury (lat = 50.3159, long = 4.0856), and Church Reef (lat = 50.3159, long = −4.0844). At each site, a single thallus was sampled using sterile forceps, transported to the laboratory at 4°C and processed within 1 h. Each of the three thalli were inspected using a dissecting microscope to remove any debris or invertebrates and separated into 1 g subsamples (N = 5). To separate the intra- and extrathalline communities, thalli were washed twice by placing in a universal tube with 20 mL sterile seawater and rotating for 10 min. Wash water was filtered through a 0.45 μm cellulose nitrate filter. Both the washed thalli (intrathalline community) and filter (extrathalline community) were homogenized using a Mini-Beadbeater machine (Biospec Products) at 42 rpm for 40 s.
DNA extraction
DNA was extracted from the washed thalli (intrathalline) and the wash water filters (extrathalline) using the PowerSoil DNA Isolation Kit (MoBio, CA, USA) and stored at −20°C. The V4 region of the bacterial 16S rRNA gene was amplified using primers 515F and 926R (Caporaso et al., Reference Caporaso, Lauber, Walters, Berg-Lyons, Lozupone, Turnbaugh, Fierer and Knight2011) using Phusion® High-Fidelity DNA Polymerase (New England Biolabs) and PCR was performed under the following conditions: 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 53°C for 30 s and 72°C for 1 min, and a final elongation step at 5°C for 10 min. The V4 region of the eukaryotic 18S gene was targeted with the primer pair E572F and E1009R (Comeau et al., Reference Comeau, Li, Tremblay, Carmack and Lovejoy2011), and PCR was performed under the following conditions: 98°C for 30 s, followed by 30 cycles at 98°C for 10 s, 55°C for 30 s, 72°C for 30 s, with a final extension at 72°C for 5 min. Sequencing was performed on the Illumina MiSeq platform (Integrated Microbiome Resource, Dalhousie University, Canada).
Bioinformatics
16S rRNA and 18S rRNA gene reads were processed through the DADA2 pipeline (Callahan et al., Reference Callahan, McMurdie, Rosen, Han, Johnson and Holmes2016), which resolves amplicon sequence variants (ASVs) at single-nucleotide resolution (Callahan et al., Reference Callahan, McMurdie and Holmes2017). Reads were trimmed and truncated to remove primers and low-quality sequences respectively. Following error estimation, paired-end reads were merged, and chimeras were removed. Taxonomic classification of resolved ASVs was performed using the RDP naïve Bayesian classifier (Wang et al., Reference Wang, Garrity, Tiedje and Cole2007) using SILVA version 132 (Quast et al., Reference Quast, Pruesse, Yilmaz, Gerken, Schweer, Yarza, Peplies and Glöckner2013) for 16S rRNA gene ASVs and PR2 version 4.12.0 (Guillou et al., Reference Guillou, Bachar, Audic, Bass, Berney, Bittner, Boutte, Burgaud, de Vargas, Decelle, Del Campo, Dolan, Dunthorn, Edvardsen, Holzmann, Kooistra, Lara, Le Bescot, Logares, Mahé, Massana, Montresor, Morard, Not, Pawlowski, Probert, Sauvadet, Siano, Stoeck, Vaulot, Zimmermann and Christen2013) for 18S rRNA gene ASVs. Read counts, taxonomic classifications, and sample metadata were compiled as a phyloseq object prior to downstream analysis (McMurdie & Holmes, Reference McMurdie and Holmes2013). L. pygmaea reads were removed from analysis. A median of 15,115 16S rRNA gene reads and 1551 processed 18S rRNA gene reads per sample were recovered (Raw sequencing data are deposited in the European Read Archive under ENA study PRJEB43698). Alpha- (Observed, Shannon Index, Pielou's Evenness) and beta-diversity (Bray–Curtis dissimilarity) analyses were carried out using phyloseq and differential abundances of specific ASVs between the extra- and intrathalline samples were assessed using DESeq2 (Love et al., Reference Love, Huber and Anders2014), based on non-normalized read count data and a significance threshold of α = 0.05, with a value of +1 assigned to the matrix to allow for statistical comparison between samples where no reads were returned. This analysis allowed for a direct test of which ASVs are enriched within the thalli and therefore most likely to be acting as photobionts. All analyses were performed in the R environment (R Core Team, 2017).
Phylogenetic trees
Identities of cyanobacterial (accession numbers MZ169867–MZ169909) and eukaryote algal (accession numbers MZ190002–MZ190020) ASVs identified by DESeq2 were confirmed by performing BLAST searches (megablast, excluding uncultured/environmental sample sequences, e-value = 0.05) against the NCBI nt database (9 November 2020) (see Supplementary material). Two 18S rRNA gene sequences (ASV_433 and ASV_251) had indeterminate identities and were removed from further analysis. To build phylogenetic trees, best BLAST hits for each ASV and an additional 2–5 close matches were obtained. Where possible, information about source material was recorded. L. pygmaea derived Rivularia sequences from Ortiz-Álvarez et al. (Reference Ortiz-Álvarez, de los Ríos, Fernández-Mendoza, Torralba-Burrial and Pérez-Ortega2015) were included in the cyanobacterial tree and photobionts of Hydropunctaria maura and Wahlenbergiella striatula from Thüs et al. (Reference Thüs, Muggia, Pérez-Ortega, Favero-Longo, Joneson, O'Brien, Nelsen, Duque-Thüs, Grube, Friedl, Brodie, Andrew, Lücking, Lutzoni and Gueidan2011) in the algal tree. Sequences were aligned using MAFFT (using G-INS-i) and manually trimmed to remove gappy tails and poorly aligned regions. The 16S rRNA gene sequence ASV_170 was poorly aligned and excluded from further analysis. Phylogenies were reconstructed using the Cipres (Miller et al., Reference Miller, Pfeiffer and Schwartz2011) implementation of RAXML-HPC (Stamatakis, Reference Stamatakis2014) using the GTR-CAT model and 1000 bootstrap replicates. Outgroups were specified for both Cyanobacteria (Pseudanabaena) and Ulvophyceae (Trentepohlia) trees. Resulting trees were first checked in FigTree (https://github.com/rambaut/figtree), before being annotated in R using ggtree (Yu et al., Reference Yu, Smith, Zhu, Guan and Lam2017). All figures were manually edited for aesthetics in Inkscape (https://inkscape.org/).
Results
Cyanobacteria dominated the bacterial community within all three thalli accounting for up to 49% of 16S rRNA gene reads (Figure 2A), while the green algae family Ulvophyceae (Chlorophyta) dominated the intrathalline eukaryotic community, accounting for up to 82% of the 18S rRNA gene reads (Figure 2A). Analyses of alpha-diversity indicated that despite increased abundance, Cyanobacteria exhibited significantly lower diversity (Shannon, P ≤ 0.001) and evenness (Pielou, P ≤ 0.001) inside the sampled thalli (Figure 2B). Similarly, Ulvophyceae showed a non-significant (Shannon, P = 0.0781) reduction in diversity within the thalli and a significant reduction in evenness (Pielou, P ≤ 0.005) (Figure 2B). Analyses of beta-diversity revealed a clear distinction between the intrathalline and extrathalline communities of both Cyanobacteria (Adonis, P ≤ 0.001) and Ulvophyceae (Adonis, P ≤ 0.001) (Figure 2C).
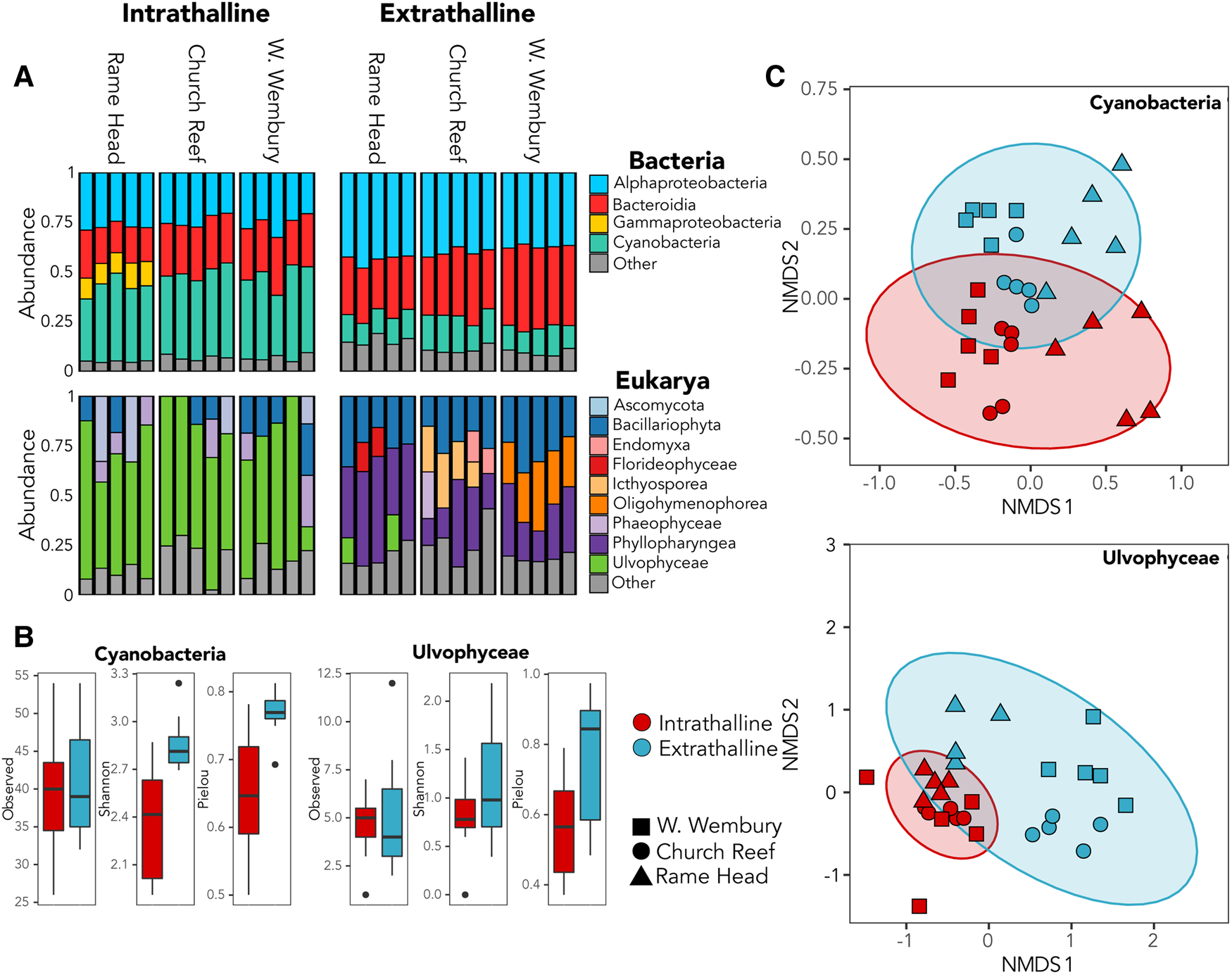
Fig. 2. (A) Relative abundance of bacterial and eukaryotic intrathalline and extrathalline communities. (B) Comparison of alpha-diversity between intrathalline (red) and extrathalline (blue) communities of Cyanobacteria and Ulvophyceae, indicating Observed diversity, Shannon Index and Pielou's Evenness. (C) Bray–Curtis NMDS plots showing beta-diversity of Cyanobacteria and Ulvophyceae intrathalline (red) and extrathalline (blue) communities. Shape indicates sampling site.
A total of six dominant (average normalized read count >20) cyanobacterial ASVs were significantly enriched (P ≤ 0.005) within at least one location (Figure 3A). Three significantly enriched ASVs, ASV_6 (MZ169869), ASV_18 (MZ169873) and ASV_55 (MZ169883) accounting for up to 8.7, 4.6 and 1.5% of the intrathalline bacterial community respectively, and a fourth ASV_73 (MZ169887), which was only found at Rame Head where it comprised up to 6.1% of the community, all belonged to Rivularia, the established L. pygmaea photobiont (Ortiz-Álvarez et al., Reference Ortiz-Álvarez, de los Ríos, Fernández-Mendoza, Torralba-Burrial and Pérez-Ortega2015). However, ASV_2 (MZ169867) and ASV_4 (MZ169868) (ASV_4 was only found at West Wembury) which made up to 20.6% and 18.1% of the intrathalline bacterial community respectively were both related to Pleurocapsa, a genus not previously associated with L. pygmaea. Other cyanobacterial ASVs were derived from the genera Acaryochloris, Phormidesmis and Synechocystis, some of which showed increased abundance in the extrathalline community. Within the phylogenetic tree (100 sequences, total alignment length = 1465 bp), all Rivularia ASVs occurred within a clade containing mostly marine and saline water derived sequences, while the highly abundant Pleurocapsa ASV_2 (MZ169867) fell within a freshwater dominated clade (Figure 4).
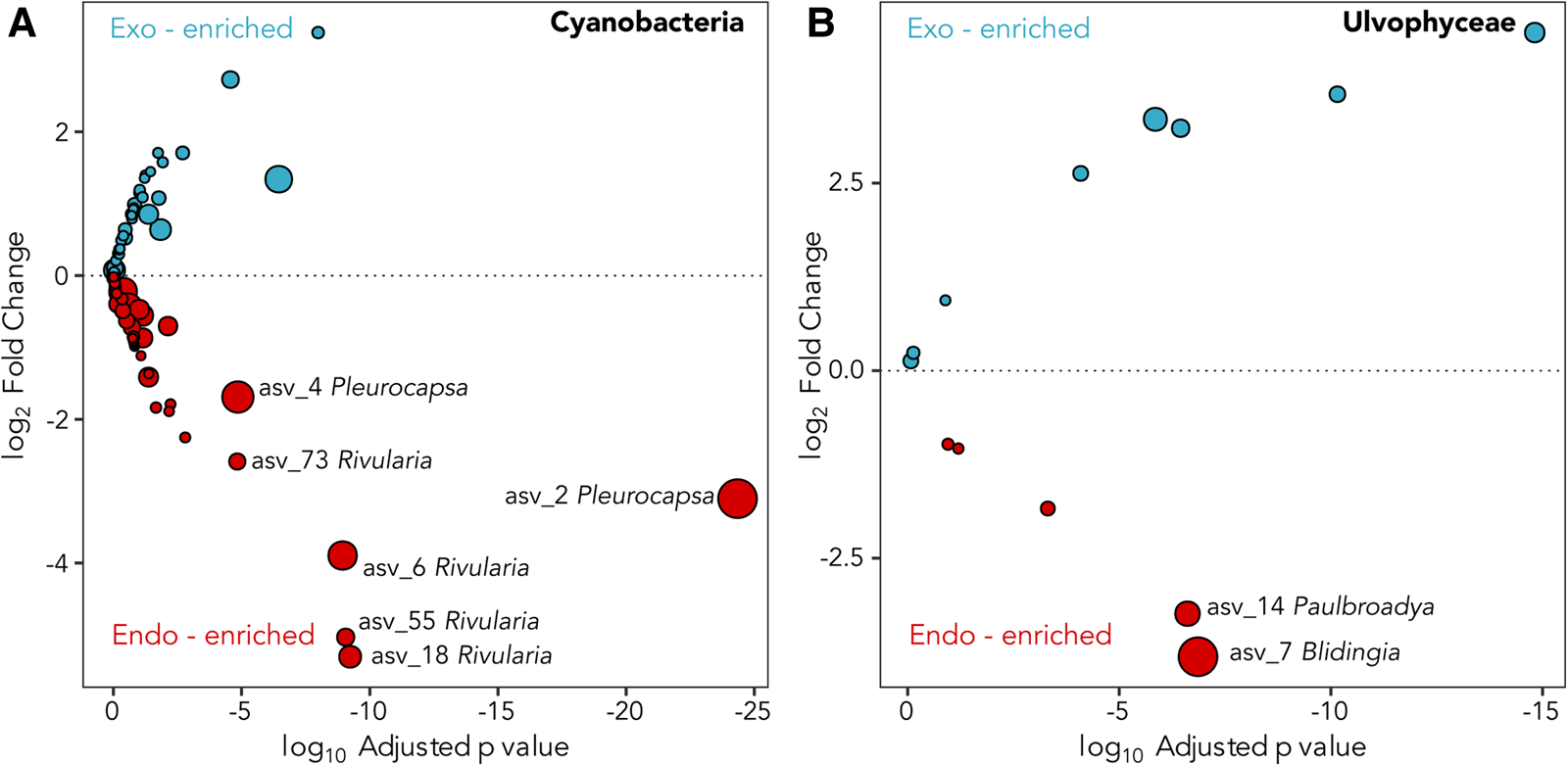
Fig. 3. Deseq2 plots indicating (A) Cyanobacteria and (B) Ulvophyceae ASVs enriched in the interior (red) and exterior (blue) of the L. pygmaea thallus. Prefix ‘endo-’ refers to intrathalline and ‘exo-’ refers to extrathalline. Circle size is the average of normalized read counts over all samples. Significant enrichments are labelled.

Fig. 4. 16S rRNA gene maximum likelihood phylogeny of Cyanobacteria associated with Lichina pygmaea. ASVs significantly enriched in the intrathalline samples are indicated with*. Prefix ‘endo-’ refers to intrathalline (red circles) and ‘exo-’ refers to extrathalline (blue circles). Circle size is relative abundance. Node labels indicate bootstrap support values for main clades.
The two abundant ASVs (average normalized read count > 20) belonging to the Ulvophyceae were significantly enriched (P ≤ 0.005) in the intrathalline community (Figure 3B), ASV_14 (MZ190003) (up to 16% intrathalline eukaryotes) and ASV_7 (MZ190002) (up to 50% intrathalline eukaryotes). Phylogenetic analysis (72 sequences, total alignment length = 1163 bp) placed these within clades of marine derived Paulbroadya and Blidingia respectively (Figure 5).
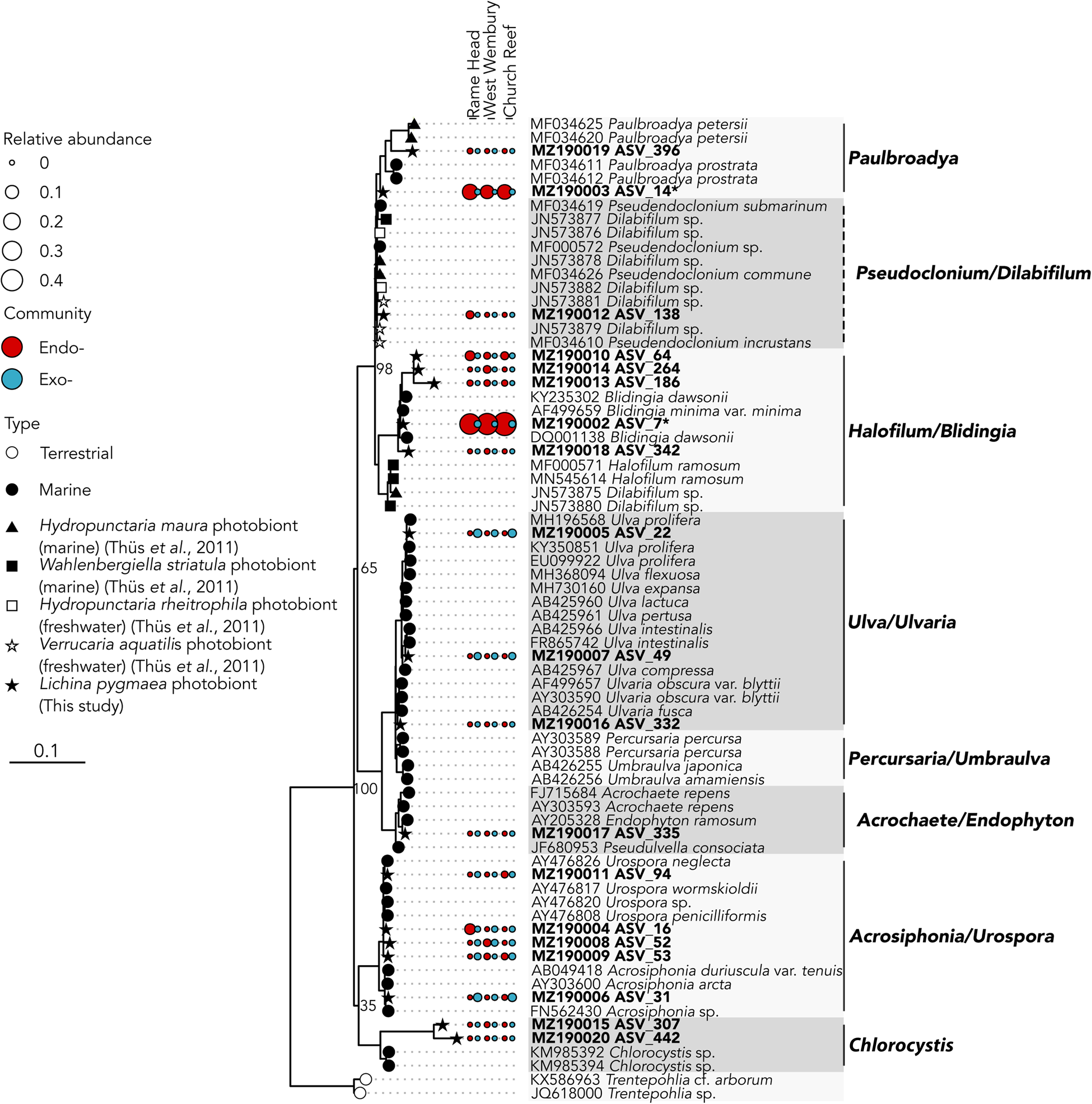
Fig. 5. 18S rRNA gene maximum likelihood phylogeny of Ulvophyceae associated with Lichina pygmaea ASVs significantly enriched in the intrathalline samples are indicated with *. Prefix ‘endo-’ refers to intrathalline (red circles) and ‘exo-’ refers to extrathalline (blue circles). Circle size is relative abundance. Node labels indicate bootstrap support values for main clades. Dashed lines next to clade names indicate paraphyletic clades.
Discussion
Here we report a considerable diversity of cyanobacterial and algal lineages associated with the interior of the L. pygmaea thallus. Complex diversity of lichen cyanobionts such as this has not been widely reported, despite early indications from the Lichinaceae family (Bubrick & Galun, Reference Bubrick and Galun1984). A previous molecular-based survey of L. pygmaea cyanobionts (Ortiz-Álvarez et al., Reference Ortiz-Álvarez, de los Ríos, Fernández-Mendoza, Torralba-Burrial and Pérez-Ortega2015), which used conventional PCR and Sanger sequencing only amplified the dominant cyanobiont Rivularia. The whole community metabarcoding approach used here (which detects all sequence variants even at low abundance) suggests that the diversity of cyanobacteria contained within the thallus of L. pygmaea may be more complex than previously considered (Ortiz-Álvarez et al., Reference Ortiz-Álvarez, de los Ríos, Fernández-Mendoza, Torralba-Burrial and Pérez-Ortega2015), and the existence of high diversity of photobionts within the lichen thallus such as shown here has likely been overlooked in many lichens. Our findings mirror those of Onuț-Bränström et al. (Reference Onuț-Brännström, Benjamin, Scofield, Heiðmarsson, Andersson, Lindström and Johannesson2018) who discovered multiple Trebouxia genotypes in Thamnolia and Cetratia thalli following Ion Torrent sequencing, compared with a single genotype recovered by Sanger sequencing. More widespread use of community metabarcoding approaches to investigate photobiont diversity may therefore reveal a greater diversity of photobionts and hitherto unrecognized lichen partnerships.
While many terrestrial tripartite lichens are well recognized (Rikkinen et al., Reference Rikkinen, Oksanen and Lohtander2002; Henskens et al., Reference Henskens, Green and Wilkins2012), to the authors' knowledge the presence of Ulvophyceae within the L. pygmaea thallus is the first documented occurrence of a putative chlorobiont in this marine species. This potentially places L. pygmaea amongst several other cyanolichens shown to also contain chlorobionts (Henskens et al., Reference Henskens, Green and Wilkins2012). In particular, L. pygmaea appears to associate with Blidingia, a blade-like multicellular alga that forms an unusual association with Turgidosculum ulvae (Verrucariaceae) whereby the mycobiont colonizes algal filaments (Pérez-Ortega et al., Reference Pérez-Ortega, Miller and Ríos2018). The exact nature of the relationship of L. pygmaea and Blidingia is yet to be established, but hints toward a unique interaction that has been previously overlooked. The other potential photobiont, Paulbroadya is known to form symbioses with several other freshwater and marine lichens in the family Verrucariaceae (Thüs et al., Reference Thüs, Muggia, Pérez-Ortega, Favero-Longo, Joneson, O'Brien, Nelsen, Duque-Thüs, Grube, Friedl, Brodie, Andrew, Lücking, Lutzoni and Gueidan2011; Gasulla et al., Reference Gasulla, Guéra, de los Ríos and Pérez-Ortega2019) (Figure 5). The relationship here between the L. pygmaea-associated Paulbroadya and the same genus associated with the crustose marine lichen H. maura indicates the potential for sharing of photobionts between marine lichens. Photobiont ‘switching’ can occur between closely and distantly related lichens occupying the same habitat (Piercey-Normore & DePriest, Reference Piercey-Normore and DePriest2001; Dal Forno et al., Reference Dal Forno, Lawrey, Sikaroodi, Gillevet, Schuettpelz and Lücking2021), offering a fitness advantage through the acquisition of locally adapted photobionts, and is likely commonplace (Yahr et al., Reference Yahr, Vilgalys and DePriest2006, Reference Yahr, Vilgalys and Depriest2004; Magain et al., Reference Magain, Miadlikowska, Goffinet, Sérusiaux and Lutzoni2017). The potential for photobiont sharing in L. pygmaea is supported by the repeated occurrence of the same Ulvophyceae ASVs between sample sites, and comparisons between photobiont communities in L. pygmaea and H. maura from the same site are now necessary to establish whether the same algal genotypes occur within both species of lichen. Interestingly, this is in contrast to several of the cyanobacterial lineages, including Rivularia, which exhibited variation between sites indicating either reduced dispersal and/or vertical transmission of cyanobionts leading to subsequent localized population structuring.
The phylogenetic placement of many of the potential L. pygmaea photobionts identified in this study within clades dominated by marine-derived lineages suggests that the lichen draws from a pool of marine photobionts, supporting the concept that photobiont selection is influenced by prevailing environmental conditions (Piercey-Normore & Deduke, Reference Piercey-Normore and Deduke2011). Although the full ecological significance of the apparent complex photobiont diversity in L. pygmaea is yet to be determined, a selection of photobionts that enhance survival under different conditions could be a key factor allowing the lichen to occupy the marine environment. Other studies have proposed that the presence of multiple coexisting photobionts with different physiological properties may contribute to increased fitness of the lichen holobiont (Yahr et al., Reference Yahr, Vilgalys and DePriest2006; Casano et al., Reference Casano, del Campo, García-Breijo, Reig-Armiñana, Gasulla, del Hoyo, Guéra and Barreno2011). Since L. pygmaea is able to photosynthesize both in air and submerged in seawater (Raven et al., Reference Raven, Johnston, Handley and McINROY1990), the ability of L. pygmaea to host a variety of photobionts may provide it with an ecological advantage with multiple marine-derived photobionts promoting survival in the intertidal zone by photosynthesizing when in seawater, while the presence of some freshwater-derived photobionts may allow for increased rates of photosynthesis at low tide and during periods of high freshwater input (e.g. rain events).
Further research into the relationship between L. pygmaea and associated photobionts is now necessary, including determining biogeographic factors in symbiont recruitment, establishing relatedness of photobionts between different marine lichen species and the extent of photobiont sharing, and photobiont localization within the thallus. Determining how much these photobionts contribute to carbon and, in the case of the cyanobacteria, nitrogen fixation will be important for understanding their contribution to biogeochemical processes, and relevant physiological experiments may be carried out using growth chambers and other experimental approaches (Henskens et al., Reference Henskens, Green and Wilkins2012). RNA studies could help to quantify photobiont activity in relation to environmental parameters including seasonal, tidal and diel cycles. This work could contribute to our understanding of the evolution of L. pygmaea and adaptation of the Lichinaceae to marine environments.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S002531542100062X.
Acknowledgements
The authors wish to thank two anonymous reviewers whose comments significantly improved the quality of the manuscript.
Financial support
NC, RA and MC were supported by the European Research Council (ERC) MYCO-CARB project (grant agreement number 772584).







