Dear Editor,
A large number of patients develop normal tissue injury following radiotherapy and this problem is becoming an important survivorship issue as the rate of curing cancer is increasing.Reference Kerns, Ostrer and Rosenstein 1 Although there is no biomarker in clinical use for identification of patients that would develop adverse effects following radiotherapy, two proteins including TGF-β1 and IL-6 have been shown to be involved in radiation-induced normal tissue toxicity and have been used as biomarkers for the prediction of radiotherapy-induced adverse effects in several independent studies. Different expressions of these proteins may partly explain the individual differences in radiotherapy toxicity observed in patients.
MicroRNAs (miRNAs) are short RNA molecules that regulate gene expression post-transcriptionally by binding to 3' or 5' untranslated region of protein coding RNAs and leading their degradation or repressing their translation.Reference Winter, Jung, Keller, Gregory and Diederichs 2 These noncoding RNA molecules tend to have noteworthy roles in a variety of cellular processes, especially in cellular responses to ionising radiation.Reference Halimi, Asghari, Sariri, Moslemi and Parsian 3 , Reference Mao, Liu, Zhang, Di and Sun 4 miRNAs contribute to the regulation of expression of TGF-β1 and IL-6 too. As there are several cross-talk between miRNAs and these proteins, different expressions of their related miRNAs in low-risk and high-risk patients would not be unexpected. Accordingly, it appear that miR-21 would increase, whereas miR-133, miR-590, let-7a, miR-187 and miR-223 would decrease in patients at risk of developing normal tissue toxicity following radiotherapy (Figure 1). Considering the point that miRNAs could release into the blood, different circulating levels of these miRNAs in low-risk and high-risk patients could be expected. This can be worthwhile for the prediction of radiotherapy-induced normal tissue injury due to the high stability of miRNAs in blood and their easy quantification by quantitative real-time PCR and other methods.
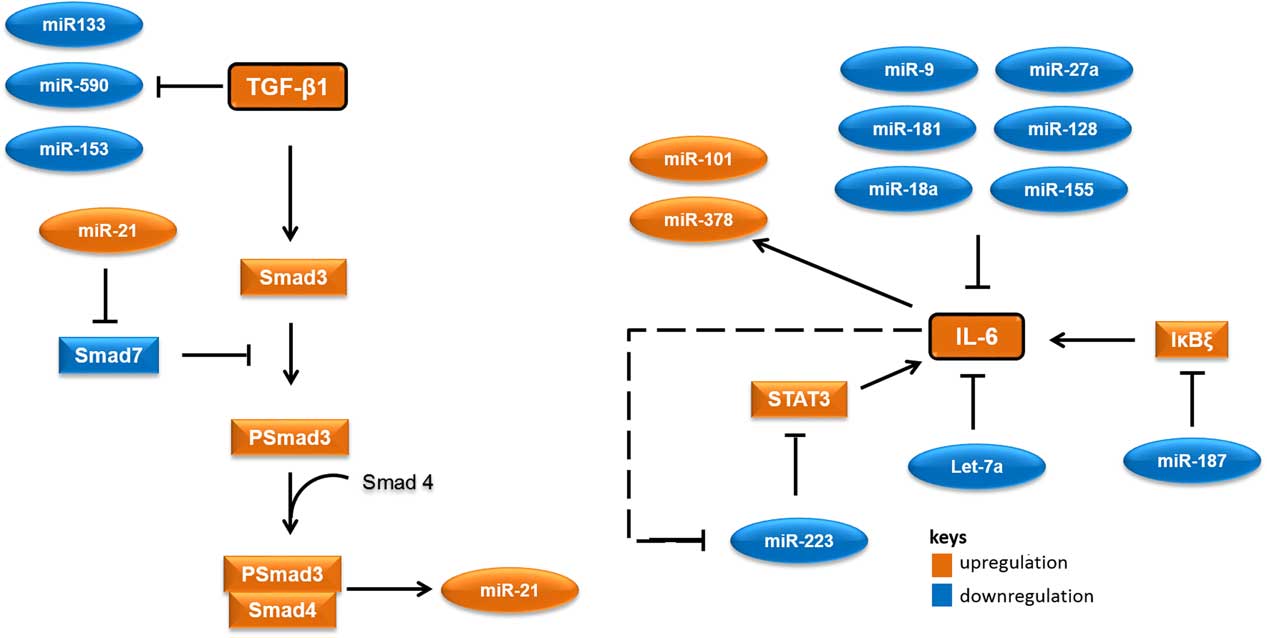
Figure 1 A simplified working model for potential microRNAs (miRNAs) involved in radiation-induced normal tissue injury and their targets. Orange colours show miRNAs and proteins that are expected to upregulate in people who would develop normal tissue injury following radiotherapy. Blue colours show miRNAs and proteins that are expected to downregulate in people who would develop normal tissue injury following radiotherapy. Abbreviations: STAT3, signal transducer and activator of transcription 3; Smad3, Mothers against decapentaplegic homolog 3 or SMAD family member 3; IκBζ, inhibitor of nuclear factor kappa B zeta; IL-6, Interleukin 6; TGF-β1, Transforming growth factor beta 1.
Sincerely,
Mohammad Halimi
Acknowledgements
This work was supported by Islamic Azad University, Babol branch.
Conflicts of Interest
None.