Introduction
The importance of microbiome in human health outcomes has been well recognised. The development and maturation of the gut microbiome occurs in early life, coinciding with the period referred to as the ‘first thousand days’.Reference Wopereis, Oozeer, Knipping, Belzer and Knol 1 This period is considered as a unique window of opportunity to promote lifelong healthy growth and development.Reference Cusick and Georgieff 2 Early exposures to microbiota, during the ‘first thousand days’, result in an expansion of these communities throughout the body whereby they occupy and/or create ecological niches that can also influence the health and wellbeing of the host.Reference Wopereis, Oozeer, Knipping, Belzer and Knol 1
Studies have demonstrated the influence of various factors and events during the first thousand days of life on the infant gut microbiota composition, including maternal health and nutrition,Reference Garcia-Mantrana and Collado 3 – Reference Macpherson, de Aguero and Ganal-Vonarburg 5 mode of delivery,Reference Madan, Hoen and Lundgren 6 – Reference Bäckhed, Roswall and Peng 9 antibiotic exposure,Reference Bokulich, Chung and Battaglia 10 – Reference Ajslev, Andersen, Gamborg, Sorensen and Jess 14 initial feeding practice (i.e. breastfeeding or formula feeding)Reference Madan, Hoen and Lundgren 6 , Reference Bäckhed, Roswall and Peng 9 , Reference Bokulich, Chung and Battaglia 10 , Reference Azad, Konya and Maughan 15 , Reference Fan, Huo and Li 16 and introduction of solid food and weaning.Reference Bäckhed, Roswall and Peng 9 , Reference Roger and McCartney 17 – Reference Koenig, Spor and Scalfone 19 Although the most rapid change in the gut microbiota occurs early in life, there is emerging evidence suggesting that the microbial community is not yet mature in adolescents.Reference Agans, Rigsbee and Kenche 20 However, most studies on early-life gut microbiota focus on the first 6 months of life. Therefore, there is a paucity of studies on the gut microbiome development and composition in late infancy, despite the critical window of the ‘first one thousand days’ extending beyond the first 6 months of life.
To fulfil the gap in understanding of the gut microbiota characteristics in late infancy, we investigated the gut microbiota in healthy 1-year-old Australian children. The aim of this study was to investigate the impact of most relevant and important factors that affect the gut microbiome during late infancy, namely mode of delivery, antibiotic exposure and breastfeeding status.
Methods
Study population and sample collection
The stool samples analysed and reported here were collected from 1-year-old children (±2 weeks) participating in the Child Health and Resident Microbes (CHaRM) study conducted in Brisbane, Australia, and before starting the multi-centre Growing Up Milk Lite (GUMLi) trial. The GUMLi trial was a double-blind recognised controlled trial to investigate the effects of growing up milk compared with regular cow’s milk in children. The inclusion criterion was otherwise healthy children turning one year of age (±2 weeks). The exclusion criteria included children who were born <32 weeks gestation; have developmental disability (i.e. autism spectrum disorder, intellectual disability); any illness or medication that may influence their nutritional status; haemoglobin concentration <100 g/l; medically diagnosed with cow’s milk allergy and inability of parents to comprehend written or spoken English.
Families participating in the GUMLi trial were invited to partake in the CHaRM study with additional consent. Ethical clearance approval was obtained from the University of Queensland Human Research Ethics Committee (reference 2014001318). Stool samples were collected by the child’s mother or caretaker in a standard faeces container (Sarstedt) and immediately home-stored at −20°C before their transfer to the laboratory on ice and long-term storage at −80°C within an average of 1.44 days (range 0–4 days). Information on duration of breastfeeding (exclusive and any breastfeeding), mode of delivery (vaginal, emergency or planned caesarean) and oral antibiotic exposure since birth, as well as other demographic and relevant data were self-reported by mothers in a face-to-face interview using a set of questionnaires. These data were collected at the time of enrolment of the study when their children were aged around 1 year.
Faecal DNA extraction and gut microbiota sequencing
The stool subsamples weighing 0.15 g were transferred to screw-capped tubes containing sterile zirconia beads (0.1 and 0.05 mm diameter). The samples were subjected to repeated bead beatingReference Yu and Morrison 21 and the released DNA was concentrated and purified using the automated Maxwell 16 MDx system (Promega, USA). The resulting DNA was used as the template to produce polymerase chain reaction (PCR) amplicons of the V3/V4 region of bacterial 16S-rRNA genes using primers (341F-CCT ACG GGN GGC WGC AG; 805R-GAC TAC HVG GGT ATC TAA TCC).Reference Klindworth, Pruesse and Schweer 22 Samples were indexed for barcoding, cleaned up and pooled before sequencing with the Illumina MiSeq platform (Illumina, USA) at the Australian Centre for Ecogenomics (Brisbane, Australia). Raw sequences were joined, demultiplexed and quality-controlled using the Quantitative Insights Into Microbial Ecology (QIIME) version 1.9.1 pipeline.Reference Caporaso, Kuczynski and Stombaugh 23 The chimera check and removal was conducted using USEARCH version 6.1.554.Reference Edgar and Flyvbjerg 24 The operational taxonomic unit (OTU) table was assigned using PyNASTReference Caporaso, Bittinger and Bushman 25 with a 97% sequence similarity threshold against the Greengenes database version 13.8.Reference DeSantis, Hugenholtz and Larsen 26 The raw OTU counts were normalized using the cumulative sum scaling method and log transformed to account for the non-normal distribution of taxa before analyses.
Statistical analyses
Demographic information is reported as the median and interquartile range (IQR), as these data were not normally distributed, except for the age of children, which was reported as mean and standard deviation (sd). To investigate the effect of selected factors on the gut microbiome, analyses were carried out between children who were grouped according to various factors whether they were still being breastfed or not, by mode of delivery and whether any antibiotics were given to the child since birth. The χ2-test was used to evaluate the differences between categorical variables. The Mann–Whitney U-test was used to compare α-diversity measures (Chao 1, Shannon diversity index, number of OTUs) based on breastfeeding status, mode of delivery and antibiotic exposure since birth. Principal component analysis (PCA) and the analysis of similarities (ANOSIM) test were used to visualise and confirm significant differences in the infant gut microbiomes. Redundancy analysis (RDA) was used to examine the community composition and cluster. The Wilcoxon rank-sum test was carried out to measure significantly different taxa between groups, where P-value of <0.05 was considered statistically significant and adjusted for multiple testing using false discovery rate (FDR). Statistical analyses of demographic information were conducted using Stata v.13.1 (StataCorp, TX, USA). Bioinformatics analyses were carried out using QIIME and Calypso.Reference Zakrzewski, Proietti and Ellis 27
Results
Description of the study cohort
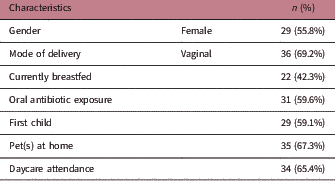
The cohort comprised 52 healthy Australian children aged 1 year (±0.03), and their general characteristics are listed in Table 1. None of the children were taking antibiotics at the time of sample collection. Two children were not born at full term (i.e. born <37 weeks gestation), but these children were corrected for gestational age at the time of sampling. The median duration of exclusive breastfeeding was 17.3 weeks (IQR 7.6–25.9), and any breastfeeding (i.e. breastfeeding alongside solids intake with or without using infant formula) was 37.9 weeks (IQR 19.5–51.9). The median duration of exclusive breastfeeding was longer for children who were still being breastfed at 1 year of age but not different for mode of delivery or antibiotic exposure. There were no significant differences in gender, birth order, pet ownership, daycare attendance, the age of solid food introduction or socio-economic status between groups of children based on current breastfeeding status, different modes of delivery or oral antibiotic exposure (data not shown).
Table 1 Characteristics of the cohort of children (n=52) aged 1 year
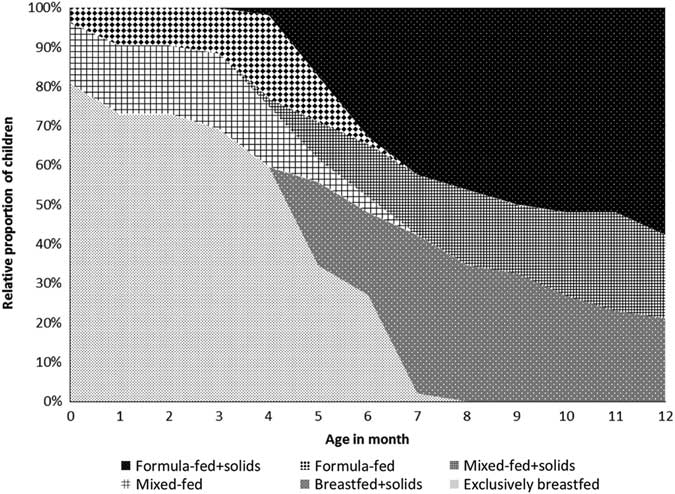
The feeding patterns since birth are illustrated in Fig. 1. During the first month after birth, 42 children (81%) were exclusively breastfed, whereas eight children (15%) were mixed-fed (having both breastmilk and formula milk) and two children (4%) were formula-fed. The median age of solid food introduction was 5 months (IQR 4–6). At the time of sampling (aged 12 months), along with solid food 11 children (21%) were breastfed, 11 children (21%) were mixed-fed and 30 children (58%) were formula-fed.
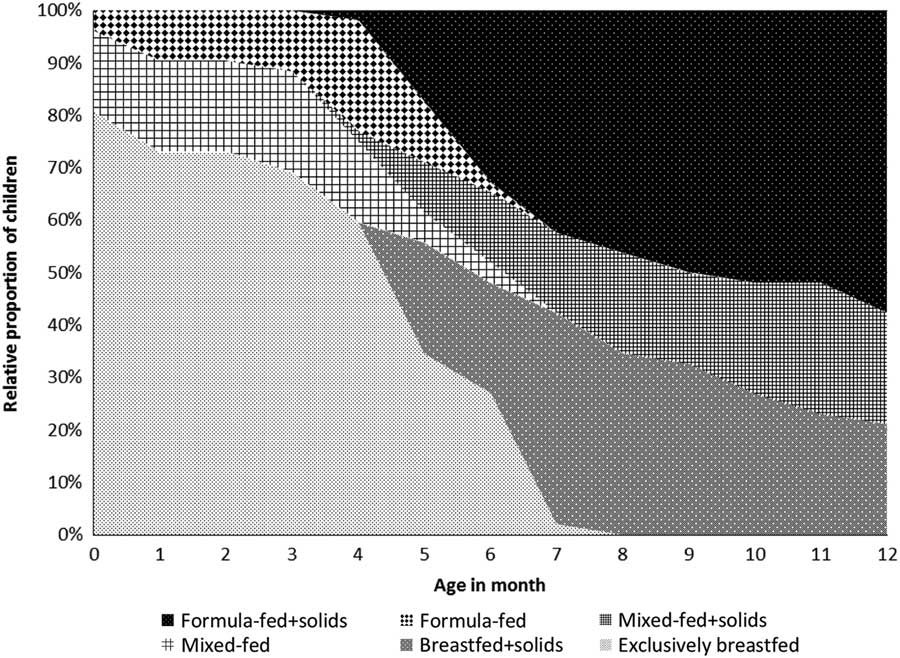
Fig. 1 Feeding patterns since birth (n=52).
Breastfeeding influences gut microbiome community structure in late infancy
The most relevant factors that modulate gut microbiota in early life, namely breastfeeding status, mode of delivery and antibiotic exposure since birth were used as constraints for the microbiota data to evaluate their impact on microbial structure variability. PCA and identification of clustering using ANOSIM were performed for all three factors. No distinct clusters were observed in PCA according to breastfeeding status, mode of delivery or by oral antibiotic usage (Supplementary Fig. 1). Breastfeeding status was the only significant variable influencing community structure based on ANOSIM (R=0.085, P<0.05) (Fig. 2). There were no significant differences in α-diversity between children based on current breastfeeding status, mode of delivery or by antibiotic exposure (Supplementary Table 1).
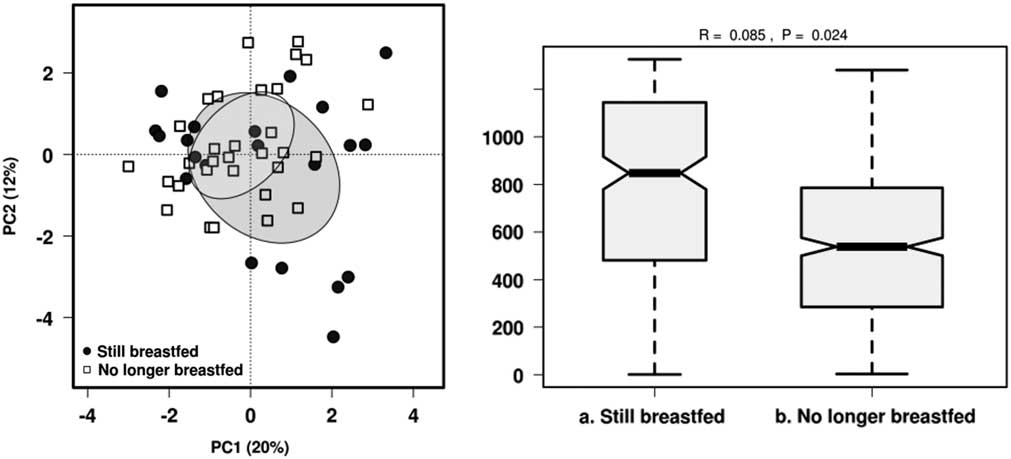
Fig. 2 Effect of breastfeeding status on the gut microbiota diversity in 1-year-old children using PCA and ANOSIM (Bray–Curtis) at genus level.
Late infancy gut microbiota taxa vary with breastfeeding status, mode of delivery and antibiotic exposure
Significantly different taxa (P<0.05) were examined at family, genus and OTU levels between currently breastfed and non-breastfed children using the Wilcoxon rank-sum analysis (Supplementary Fig. 2). At the family level, Veillonellaceae was higher among the continued breastfeeding group (FDR 0.072), whereas unclassified Clostridiales was higher among children who were no longer being breastfed (FDR 0.072). At the genus level, Veillonella was higher among continued breastfeeding group (FDR 0.042). At the OTU level, Veillonella dispar (OTU757622) was higher in the continued breastfeeding group but not after adjusting for multiple testing (FDR 0.65). Amongst the group of children who were no longer breastfed, Barnesiellaceae OTU315846, Clostridiaceae OTU555945, Ruminococcaceae OTU309720, Blautia OTU583089 and Clostridiales OTU558444 were higher but not after adjusting for multiple testing (FDR 0.65). There were no significant differences in bacterial taxa based on mode of delivery or exposure to oral antibiotics after multiple testing adjustment.
Different feeding patterns further differentiate gut microbiome community structure in late infancy
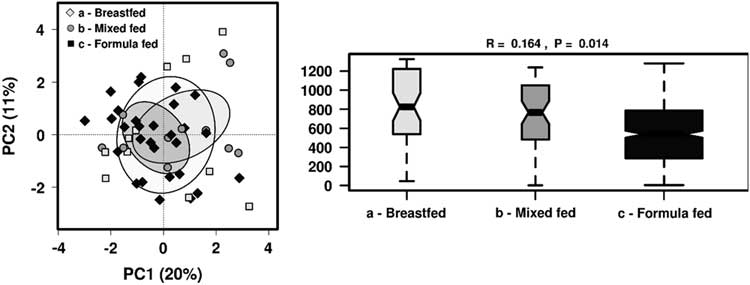
As we found that breastfeeding status showed most significant differences in the gut microbiome taxa profile, we further analysed the effect of feeding patterns by dividing children into groups of those who were breastfed, mixed-fed or formulate-fed alongside solid food intake at the time of sample collection . There were no distinct clusters observed in PCA but ANOSIM showed feeding patterns significantly influenced the gut microbiome community structure (R=0.164, P=0.014) (Fig. 3). Significantly different taxa (P<0.05) were investigated among these groups, as well as to identify any significant clustering by RDA (Fig. 4).
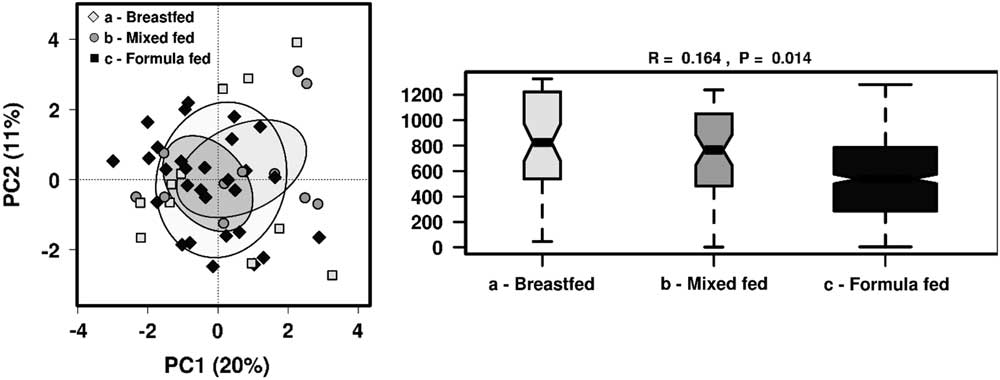
Fig. 3 The effect of different feeding patterns on the gut microbiota diversity in 1-year-old children using PCA and ANOSIM (Bray–Curtis) at genus level.
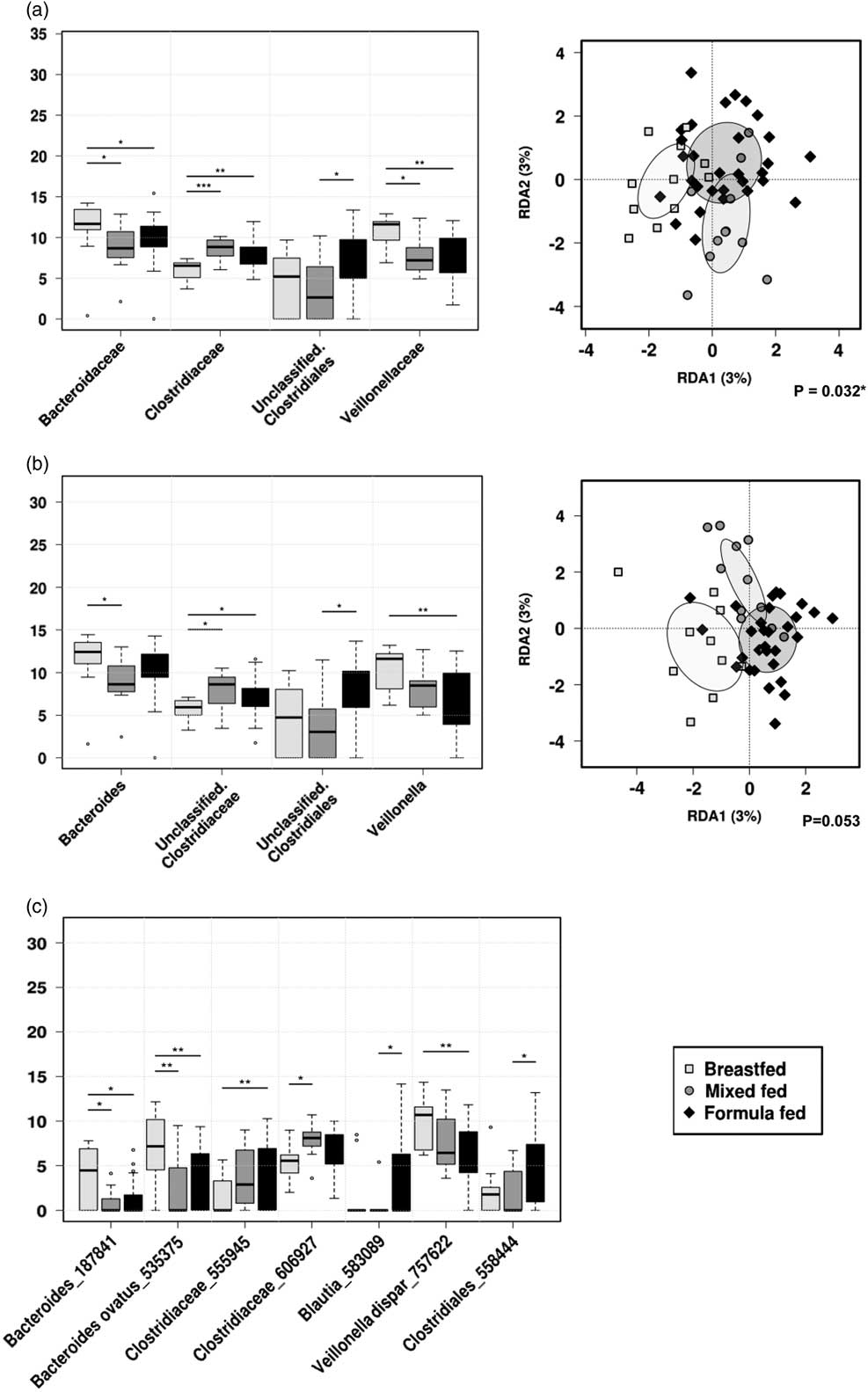
Fig. 4 Rank-sum box plots showing the different taxa and RDA between groups of children who were receiving breastmilk only, mixed-fed or formula milk alongside solid food at 1 year of age at (a) family; (b) genus; (c) OTU levels. *P<0.05, **P<0.01, ***P<0.001 (RDA analysis at OTU level not significant).
At the family level, Veillonellaceae (FDR 0.032) and Bacteroideceae (FDR 0.16) were significantly higher amongst the breastfed group compared with mixed-fed or formula-fed groups, whereas Clostridiaceae was significantly higher amongst mixed-fed group compared with breastfed or formula-fed groups (FDR 0.032). Unclassified Clostridiales were higher among the formula-fed group compared with the mixed-fed group (FDR 0.16). At the genus level, Veillonella was significantly higher in the breastfed group compared with both mixed-fed and formula-fed groups (FDR 0.067), and Bacteroides was higher in the breastfed group compared with mixed-fed group (FDR 0.33). Unclassified Clostridiaceae was higher in the mixed-fed group compared with breastfed or mixed-fed groups (FDR 0.33), and an unclassified Clostridial was significantly higher in the formula-fed compared with the mixed-fed group (FDR 0.33).
Examination of the microbial community structure between the three groups by the hypothesis-based RDA analysis showed that different feeding patterns were significantly associated with the gut microbiota composition at the family level (P<0.05) in 1-year-old children.
Other possible antibiotic exposure, different caesarean delivery or prematurity did not alter gut microbiome community structure in late infancy
As the data were collected retrospectively, details of the antibiotic exposure could not be verified. Some children may have been exposed to prophylactic antibiotics during perinatal period from caesarean delivery. Furthermore, evidence exists for a different microbial community profile between children born by emergency or planned caesarean. Table 2 shows the distribution of emergency and caesarean delivery, as well as oral antibiotic exposure and all antibiotic exposure. We first explored differences in gut microbiota profiles between children born by emergency and planned caesarean section delivery but found no significant difference in the microbial community structure between the groups. Next, a stratified analysis of all children who may have been exposed to antibiotics (n=39) was performed. Again, breastfeeding status (R=0.149, P<0.05) and different feeding patterns (R=0.241, P<0.05) showed significant influence on the community structure based on ANOSIM (Supplementary Fig. 3). To examine the effect of prematurity, further sub-analysis was conducted by excluding prematurely born infants (n=2). This did not alter the significance of the finding, where the breastfeeding status remained as the only significant factor influencing the microbial community status.
Table 2 Distribution of different modes of delivery and oral antibiotic exposure among children currently breastfed and no longer breastfed at age 1 year
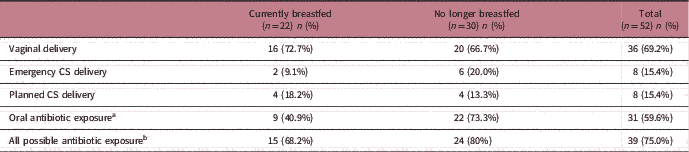
CS, caesarean section.
a Oral antibiotic exposure regardless of mode of delivery.
b Children exposed to either or both oral antibiotics and/or caesarean delivery.
Breastfeeding may attenuate the effect of antibiotic exposure on the gut microbial community
To further test the effect of breastfeeding, we ran stratified analyses of children who were still being breastfed and those who were no longer breastfed. In the analysis of children who were still being breastfed (n=22), we did not find any significant difference with the mode of delivery or antibiotic exposures. Although there were no microbial community structural differences identified using ANOSIM among children who were no longer being breastfed (n=30), we found that oral antibiotic exposure was associated with higher Oscillospira (P<0.01, FDR 0.17) and SMB53 (P<0.05, FDR 0.38) (Supplementary Fig. 4). For all possible antibiotic exposure (i.e. oral antibiotics and born by caesarean delivery), Oscillospira was again higher (P<0.01, FDR 0.28). Other taxonomical differences were not significant after adjusting for multiple corrections (FDR>0.5).
Discussion
This study showed that breastfeeding had significant influence on the microbial community structure in this healthy cohort of 1-year-old children, whereas mode of delivery or exposure to antibiotics since birth had minor structural effects on the gut microbiota in late infancy. Although the value was small, breastfeeding status and feeding mode showed statistically significant differences in community profiles. In the broad analysis of continued breastfeeding, breastfed children harboured more lactate utilisers, whereas children who were no longer breastfed were enhanced with butyrate producers. The microbial family that was most consistently associated with breastfeeding status was Veillonellaceae in this present study, after correcting for multiple testing.
Large amounts of lactate are produced from undigested substrates, including human milk oligosaccharides (HMOs), thereby requiring lactate utilising bacteria such as Veillonella to prevent lactate accumulation by converting lactate to short-chain fatty acids (SCFAs), acetate and propionate.Reference Duncan, Louis and Flint 28 – Reference Pham, Lacroix, Braegger and Chassard 30 Breastfed children have higher satiety response than formula-fed children,Reference Brown and Lee 31 which could be partially explained by different SCFAs produced by the gut microbiota. Propionate is associated with satiety and improvements in blood glucose responsivenessReference Arora, Sharma and Frost 32 and reduction in food intake.Reference Lin, Frassetto and Kowalik 33 The measurement of SCFA would be advantageous to confirm these results.
Our finding is that Veillonellaceae was significantly higher among those children who were only having breast milk alongside solid food, compared with mixed-fed children, who were having both breast milk and formula milk, suggesting that Veillonellaceae may be a poor substrate competitor leading to its reduction when breastfeeding is reduced. This has been supported by a metagenomics study of infant gut microbiota, which also identified that cessation of breastfeeding shifted the microbiota population towards a more adult-like profile.Reference Bäckhed, Roswall and Peng 9 Veillonella has been identified as a common species between the maternal gut, oral and placenta microbiota,Reference Gomez-Arango, Barrett and McIntyre 34 associated with an inherent maternal gut microbiomeReference Bäckhed, Roswall and Peng 9 and is commonly found in neonates. There are also other reported positive associations with Veillonella in early-life health status. For instance, genus Veillonella is negatively associated with the likelihood of bronchiolitis,Reference Hasegawa, Linnemann and Mansbach 35 and the reduction of its relative abundance has been significantly linked in children at risk of asthma.Reference Arrieta, Stiemsma and Dimitriu 36 Thus, Veillonella spp. may have impact on immunomodulatory effects that are beneficial during the early-life developmental period.
Continued breastfeeding appears to delay progression of specific bacterial taxa, such as butyrate producers. We found that order Clostridiales and OTUs belonging to Clostridiaceae and Ruminococcaceae, all belonging to Firmicutes phylum, were higher among groups of children who were no longer breastfed and those who were formula-fed or mixed-fed, suggesting increased population of butyrate producers from consumption of solid food and/or infant formula. It was previously reported that after the introduction of solid food, Clostridiales and Blautia increased significantly among infants who were having milk other than breastmilk,Reference Thompson, Monteagudo-Mera, Cadenas, Lampl and Azcarate-Peril 37 which is in line with our finding.
Generally, breastfed infants have the gut microbiota profile dominated by Bifidobacterium.Reference Harmsen, Wildeboer-Veloo and Raangs 38 , Reference Jost, Lacroix, Braegger and Chassard 39 However, we did not find significant differences in the abundance of Bifidobacterium according to breastfeeding status. Bifidobacterium are known for its HMO utilising propertiesReference Garrido, Dallas and Mills 40 and health-enhancing benefits.Reference Di Gioia, Aloisio, Mazzola and Biavati 41 It may be possible that prebiotics in infant formula, as well as fibre from other dietary sources may have contributed to undifferentiated levels of Bifidobacterium amongst this cohort of healthy children.Reference Bakker-Zierikzee, Alles and Knol 42 – Reference Scholtens, Alles and Bindels 44
However, we found that Bacteroides ovatus OTU535375 was significantly higher species associated with breastfeeding. Bacteroides species have the capacity to degrade different types of polysaccharides,Reference Marcobal, Barboza and Sonnenburg 45 which give them competitive advantage over other bacterial groups that do not have this capacity.Reference Nylund, Satokari, Salminen and de Vos 46 The role of Bacteroides in the breakdown of HMO was demonstrated in a previous metagenomics-based study, which found Bacteroides species as dominant contributor to HMO breakdown.Reference Yassour, Vatanen and Siljander 11 Therefore, Bacteroides species are one of the first bacterial groups to colonise a breastfed infant’s gutReference Palmer, Bik, DiGiulio, Relman and Brown 47 and continues its presence as one of the major core gut microbiota throughout life.Reference Scholtens, Oozeer, Martin, Amor and Knol 48 , Reference Huse, Ye, Zhou and Fodor 49
Another key finding of this study is that neither mode of delivery nor did antibiotic exposure result in differences in gut microbiota richness or structure in this cohort of this children. Yassour et al.Reference Yassour, Vatanen and Siljander 11 found that when children were exposed to antibiotics during their first year of life, faecal microbiota richness was reduced compared with those not exposed to antibiotics, but this was only apparent in samples collected after the first year of life. Therefore, children in this current study may experience changes in microbial communities in the subsequent period. However, differences in microbial taxa were identified in the stratified analysis amongst children who were no longer breastfed, and we found that antibiotic exposure was associated with higher Oscillospira. This bacterial genus is generally linked with health and leanness in both children and adults;Reference Konikoff and Gophna 50 therefore, the reason behind increased Oscillospira among the antibiotic exposed children in this study needs to be further investigated. It could be speculated that not finding difference in the gut microbial community from antibiotic exposure among children who were still being breastfed suggests that breastfeeding may attenuate the effect of antibiotic exposure. Again, further study with a larger sample and more detailed information on antibiotics is needed. With regards to mode of delivery, a recent systematic review has also identified that significant differences in the diversity and colonisation pattern of the gut microbiota due to delivery mode diminish after 6 months of life,Reference Rutayisire, Huang, Liu and Tao 51 which may explain the results from our current study.
Limitations
There are limitations in this study. First, the size of the cohort, in relative terms is quiet small for observational studies. As such, it is not possible to consider how variations in host genetics alter the gut microbiome,Reference Lim, You and Yoon 52 as well as those interactions with diet that further influence the gut microbiome, leading to potential modulation of individual risk for metabolic syndromes.Reference Carmody Rachel, Gerber Georg and Luevano Jr Jesus 53 , Reference Ussar, Griffin and Bezy 54
Secondly, a detailed analysis of diet during infancy is needed to comprehensively understand the complex interactions between breastmilk, formula milk, diet and the microbiome. Furthermore, retrospective data collection did not allow detailed analysis of the exact type of antibiotic and duration of the antibiotic course given to each child in this study. However, stratified analysis of all children possibly exposed to antibiotics since birth by combining children who had oral antibiotic exposure and those who were born by caesarean delivery to account for prophylactic antibiotic use during delivery found breastfeeding status and feeding pattern remained as significant factors that differentiated the gut microbiota profile. Although this study did not identify that antibiotics exposure altered the gut microbial community in this cohort of children, this finding is in contrast with studies presented by others.Reference Bokulich, Chung and Battaglia 10 , Reference Yassour, Vatanen and Siljander 11 It is possible that early-life antibiotics exposure may have later health impact on this cohort of children. Follow-up studies would be needed to monitor such effect.
Thirdly, the importance of SCFA in health outcomes is well-established, but conditions such as pH and storage can affect the SCFA extraction yield.Reference Garcia-Villalba, Gimenez-Bastida and Garcia-Conesa 55 As the measurement of SCFA was not part of the original study protocol, and samples were stored for microbiome DNA sequencing only, SCFAs were not measured in this study. Future studies should incorporate experimental protocols that accommodate analysis of SCFA from faecal samples.
Conclusions
This study has contributed to research in the gut microbiome of healthy 1-year-old children, a group which receives considerably less focus than young infants. Previous studies found that cessation of breastfeeding was the major driver in the gut microbiota development;Reference Bäckhed, Roswall and Peng 9 , Reference Bergstrom, Skov and Bahl 56 however, we found gut microbiota characteristics amongst children who were having breastmilk and solid food, compared with those who were having both breastmilk and formula milk alongside solid food, were also different. This suggests the duration of breastfeeding, as well as overall diet enrich keystone species that could have important immune and metabolic roles during this critical developmental window. Further evaluation of the clinical relevance and physiological significance of differences in gut microbiome characteristics during this period would assist in further understanding the role of gut microbiome on long-term health outcomes.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/S2040174418000624
Acknowledgements
We would like to thank Morrison laboratory group for their support.
Financial Support
M.M. was supported by Australian Government’s Research and Training Program scholarship. The study was partially funded by Nutricia Australia Pty Ltd.
Conflicts of Interest
None.
Ethical Standards
This study received ethical clearance approval from the University of Queensland Human Research Ethics Committee (reference 2014001318).