Mastitis, particularly caused by Staphylococcus aureus is considered to be one of the most prevalent infectious maladies affecting the dairy industry which leads to severe financial losses in terms of decreased milk yield and quality (Muzammil et al., Reference Muzammil, Ijaz, Saleem and Ali2022). The emergence of antibiotic resistance in S. aureus has led to limited treatment options and number of resistant strains like methicillin-resistant S. aureus (MRSA) has evolved at the animal-human interface (Muzammil et al., Reference Muzammil, Ijaz, Saleem and Ali2022). S. aureus can produce more than one leukotoxin including Panton–Valentine leukocidin (PVL) that inhibits the immune response, allowing the microorganisms to survive and the inflammatory process to continue in soft tissues (and elsewhere). PVL toxin has been found in the isolates from methicillin-resistant and methicillin-sensitive (MSSA) S. aureus of various habitats which include humans (clinical or community-associated isolates), farm animals and animal-derived products (Abboud et al., Reference Abboud, Galuppo, Tolone, Vitale, Puleio, Osman, Loria and Hamze2021). This study was planned to estimate the prevalence and antimicrobial susceptibility of S. aureus harboring virulent genes and methicillin resistance genes responsible for mastitis in cattle of Punjab province, Pakistan.
Materials and methods
A total of 690 bovine milk samples from smallholder (n = 149), commercial (n = 284) and corporate (n = 257) dairy farms (49 farms in total) in the province of Punjab, Pakistan were collected aseptically to detect subclinical and clinical mastitis. Standardized microbiological procedures were carried out for the isolation and confirmation of S. aureus isolates. The phenotypic and genotypic confirmation of methicillin resistance was confirmed by guidelines of (Muzammil et al., Reference Muzammil, Ijaz, Saleem and Ali2022). The presence of the pvl gene in all confirmed S. aureus isolates was done using the following primer for PVL gene (luk-PV-1, ATCATTAGGTAAAATGTCTGGACATGATCCA and, for luk-PV-2, GCATCAATGTATTGGATAGCAAAAGC: Shrivastava et al., Reference Shrivastava, Sharma, Shrivastav, Nayak and Rai2018). The processing of PCR products was done by multiple sequence alignment by BioEdit software through Clustal W multiple alignment to compare the sequence of our isolates with the other highly similar published sequences from different countries. A phylogenetic tree was then constructed via maximum likelihood method at 1000 replications bootstrapping technique on MEGA-X software. The sequence of isolates connected by the same internal nodes indicates more closeness as compared to sequences connected by different internal nodes. The in vitro susceptibility profiling and prevalence of pvl positive and negative MRSA against various antibiotics was also performed, as described by Muzammil et al. (Reference Muzammil, Ijaz, Saleem and Ali2022).
Results
The data presented in Table 1 reveal an overall total of 175 (25.36%) S. aureus isolates from 690 milk samples (subclinical n = 512; clinical mastitis = 172). The prevalence of mecA and pvl gene in S. aureus isolates was recorded to be 22.29% and 6.29% respectively, while 133 isolates didn't contain either of these genes. We found the presence of both mecA and pvl gene in 8 isolates (4.57%), which we labeled as pvl positive MRSA. The phylogenetic tree analysis via maximum likelihood method at 1000 replication bootstrapping technique revealed that study sequences are more similar with each other as compared to other country's sequences. The bootstrap results showed that sequences from Egypt (FJ821791) and India (MK975993) are comparatively more related to our sequences than other published sequences. In particular, isolates from USA (EF571841), Turkey (FJ895585), Myanmar (MK902786) and China (AB678715) lie in different clades from our isolates whilst isolates from France (EU518770), UK (HM584704) and Japan (AB256039) form out-group showing least similarity with our data (Fig. 1).
Table 1. Gene pattern of mecA and pvl in S. aureus isolated from clinical and subclinical mastitis
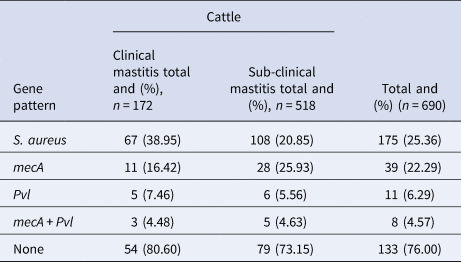
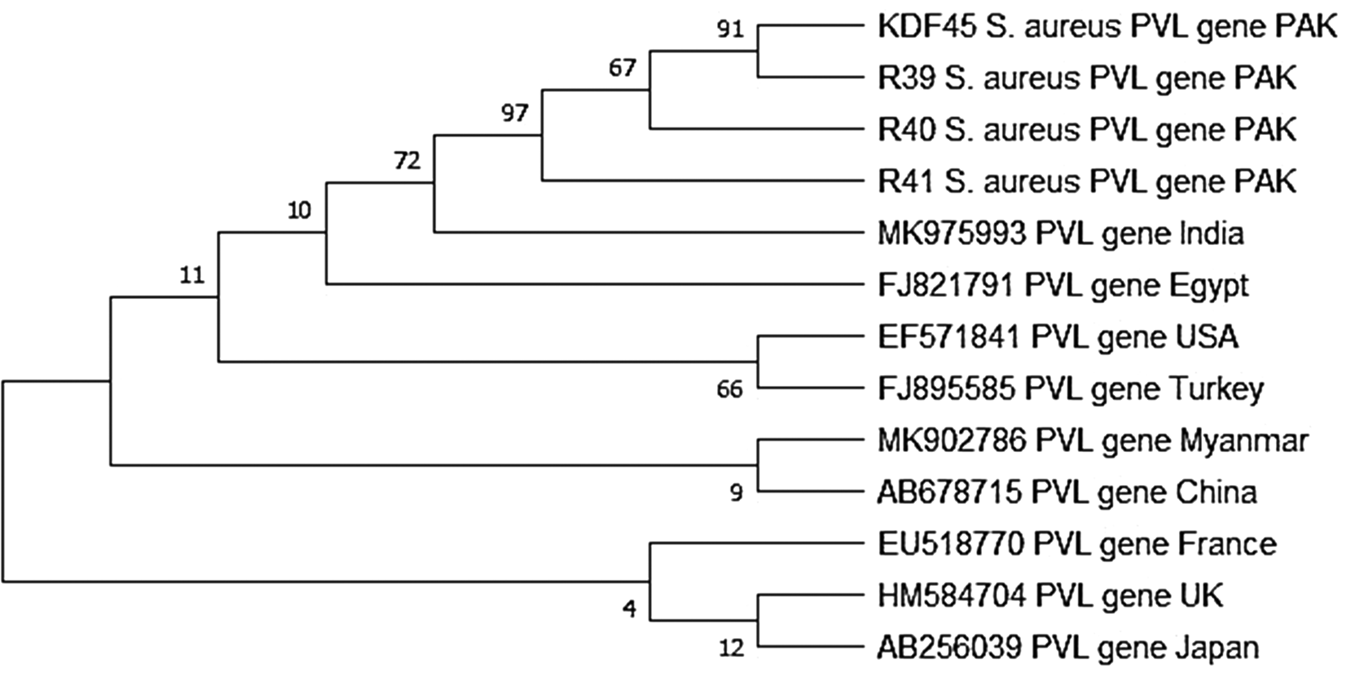
Figure. 1. Phylogenetic tree construction by 1000 bootstrapping replication indicating the relationship of local PVL isolates (PAK) with already reported sequences.
All of the 8 pvl positive MRSA isolates (Group A) showed 100% resistance towards cefoxitin, oxacillin, and vancomycin while having complete susceptibility against gentamicin, ciprofloxacin, levofloxacin, moxifloxacin, oxytetracycline, tylosin, and linezolid groups. In comparison, the pvl negative MRSA isolates (i.e. positive for mecA: Group B) also exhibited 100% resistance against cefoxitin and oxacillin but 100% susceptibility against gentamicin, tylosin, and linezolid (Table 2).
Table 2. In vitro antibiogram profile of different antibiotic groups against pvl positive and negative MRSA

R, resistant; I, intermediate; S, sensitive.
Discussion
The detected prevalence of subclinical and clinical mastitis was reported to be 75.07% and 24.93% in milk samples of cattle which differs from the earlier report of Javed et al. (Reference Javed, Ijaz, Fatima, Anjum, Aqib, Ali, Rehman, Ahmed and Ghaffar2021). The increased occurrence of subclinical mastitis in dairy livestock is due to multiple predisposing factors like contamination during milking, improper housing and improper disease management of animal (Javed et al., Reference Javed, Ijaz, Fatima, Anjum, Aqib, Ali, Rehman, Ahmed and Ghaffar2021) and in a similar vein the transmission of S. aureus between animals occurs with the use of contaminated milk utensils, improper milking hygiene and contaminated milker's hands (Schnitt and Tenhagen, Reference Schnitt and Tenhagen2020). Studies from Pakistan show that the most common agent causing mastitis is Staphylococcus spp. followed by Streptococcus spp. (Javed et al., Reference Javed, Ijaz, Fatima, Anjum, Aqib, Ali, Rehman, Ahmed and Ghaffar2021) . The prevalence of S. aureus that we recovered from subclinical and clinical mastitis cases was 20.85 and 38.95% respectively, higher than the 15.2% prevalence reported by Shrestha et al. (Reference Shrestha, Bhattarai, Luitel, Karki and Basnet2021).
Our antimicrobial susceptibility testing of S. aureus strains showed 100% resistance to cefoxitin and oxacillin, while 22.28% of S. aureus isolates were found mecA positive (only). This is much higher than the 6.9% reported by Shrestha et al. (Reference Shrestha, Bhattarai, Luitel, Karki and Basnet2021). Methicillin resistance is linked with the existence of the mecA gene on the S. aureus chromosome, which is responsible for the synthesis of penicillin-binding protein (PBP2a: Muzammil et al., Reference Muzammil, Ijaz, Saleem and Ali2022). We observed the presence of the pvl gene (alone) in 6.28% of S. aureus isolates.
The pvl gene has been identified in 10.5% of S. aureus isolates in India (Shrivastava et al., Reference Shrivastava, Sharma, Shrivastav, Nayak and Rai2018) and 41.5% of S. aureus isolates in China (Wang et al., Reference Wang, Wang, Yan, Wu, Ali, Li, Lv and Han2015). Conversely, some studies could not identify the pvl gene in any S. aureus isolates from cattle milk (Patel et al., Reference Patel, Godden, Royster, Crooker, Johnson, Smith and Sreevatsan2021). The pvl gene is known to be the most powerful staphylococcal leukotoxin that can resist bovine neutrophils (Algammal et al., Reference Algammal, Enany, El-Tarabili, Ghobashy and Helmy2020). Hence, pvl may produce resistance by attacking the bovine polymorphonuclear cells and increase pathogenicity against the host (Hata et al., Reference Hata, Katsuda, Kobayashi, Uchida, Tanaka and Eguchi2010). Our study is the first to determine the occurrence of mecA and pvl gene in S. aureus isolates from cattle milk in Pakistan.
In conclusion, we have demonstrated the presence of mecA and pvl gene in 22.29% and 6.29% of S. aureus isolates, and the combination in 4.57% of milk samples from bovine mastitis in Punjab. The presence of Panton–Valentine leukocidin toxin and methicillin resistance in mastitis-associated S. aureus poses a zoonotic threat, requiring awareness and control measures. Antimicrobial susceptibility testing is crucial for effective treatment and to prevent antimicrobial resistance problems locally in Punjab and globally.
Acknowledgement
The authors are thankful to the Punjab Higher Education Commission (PHEC) for providing funds for the current study under project no. PHEC/ARA/PIRCA 20150/4.





