Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).Reference Helmy, Fawzy, Elaswad, Sobieh, Kenney and Shehata1 Healthcare workers (HCWs) account for a large proportion of infected individuals.2 The criteria to allow infected HCWs to return to work vary between jurisdictions, and the exact duration of contagiousness remains uncertain.Reference Arons, Hatfield and Reddy3–Reference Liu, Chang and Wang6 Two main return-to-work approaches exist: symptom-based strategies and test-based strategies.7–9 Symptom-based strategies allow return to work when ≥7–14 days have elapsed since symptom onset and symptoms have resolved.7,8 Test-based strategies require work exclusion until negative results of a SARS-CoV-2 reverse-transcriptase polymerase chain reaction (RT-PCR) assay are obtained from ≥1 respiratory specimens.8,9
Both approaches use surrogate markers of loss of contagiousness (ie, symptomatology and RT-PCR). The detection of virus by culture evaluates the viral replicating capacity and is a better indicator of contagiousness. The few studies that have investigated SARS-CoV-2 infectivity using culture have found that infectivity may be resolved within 10 days.Reference Arons, Hatfield and Reddy3–Reference Bullard, Dust and Funk5,Reference Folgueira, Luczkowiak, Lasala, Perez-Rivilla and Delgado10 However, the significance of their findings is hampered by methodological issues limiting culture sensitivity. We sought to determine the presence of infective SARS-CoV-2 in HCWs with COVID-19, 2 weeks after symptom onset, using viral culture as a marker of infectivity.
Methods
Population and setting
We performed a prospective cross-sectional study in 3 academic healthcare institutions in Quebec, Canada. The provincial strategy at the time of the study allowed return to work if HCWs fulfilled the following criteria: (1) resolution of symptoms (with the exception of anosmia or residual cough); (2) ≥14 days elapsed since symptom onset; and (3) 2 consecutive respiratory specimens negative for SARS-CoV-2 by RT-PCR. The population under study included HCWs with confirmed COVID-19 who underwent control RT-PCR for tentative return to work and who met the first 2 criteria.
Nasopharyngeal samples were collected using a flocked swab (FLOQSwabs, Copan Italia, Brescia, Italy), placed in 3 mL universal viral transport media (UTM, Copan Italia) and were sent to the local microbiology laboratory within 4 hours of collection. On the same day, a 1-mL aliquot was sent to the Laboratoire de Santé Publique du Québec (LSPQ), where it was kept at 4°C and was inoculated within 24 hours.
Viral culture
Viral cultures were performed on Vero E6 cells in 4-mL shell vials. Cells were grown in Dulbecco’s modified Eagle medium (DMEM; Life Technologies, Carlsbad, CA) supplemented with L-glutamine, gentamicin, vancomycin, amphotericin B, and 10% fetal bovine serum (Hi FBS; Gibco, Carlsbad, CA) until confluence. Each culture series was controlled with a negative and a positive sample. A 0.1-mL aliquot of specimen was used as an inoculum. A negative cellular control was added daily with newly inoculated specimens. Cultures were kept for 10–15 days. The presence of a cytopathic effect (CPE) was monitored every other day.
RT-PCR
In-house RT-PCR for SARS-CoV-2 targeting the N gene were performed on the original specimens and on culture supernatants at the end of the incubation period to document either viral replication without CPE or to validate the etiology of a CPE. Nucleic acids were extracted from 0.2-mL aliquots using the easyMAG or EMag (bioMérieux, Marcy-lʼÉtoile, France). Forward, reverse, and probe sequences were as follows: AACCAGAATGGAGAACGCAGTG, CGGTGAACCAAGACGCAGTATTAT and CGATCAAAACAACGTCGGCCCCAAGGTTTAC. Assays were performed using the TaqPath One STEP RT-PCR kit on a QuantStudio3 apparatus (ThermoFisher Scientific, Waltham, MA). The N assay is routinely used to confirm results obtained with the Corman E gene assay, and it is at least as sensitive as the latter (data not shown).
The study was approved by local research ethics committees.
Results
Overall, 118 participants were enrolled between May 2 and June 2, 2020 (79% female; average age, 40 years old). The median elapsed time between symptom onset and sampling was 14 days (range, 12–24 days); 87% of samples were collected on day 14, 15, or 16. Most samples (84 of 118, 71%) were positive by RT-PCR, with a median cycle threshold (Ct) value of 32.6 (interquartile range, 30–34.5) (Fig. 1). Overall, 14 samples showed a CPE on culture. In most of these (13, 93%), RT-PCR of the supernatant was negative. However, 1 sample (1 of 118, 0.85%; 95% confidence interval, 0.02%–4.6%) exhibited a CPE on day 11 of incubation, for which RT-PCR of supernatant was strongly positive with a Ct value lower than the original specimen (Ct, 9.9 vs 34, respectively), indicating active SARS-CoV-2 replication. RT-PCR of supernatant on all samples with no CPE (n = 104) were either negative (n = 91) or had Ct values that were equivalent to or higher than the original sample (n = 13).
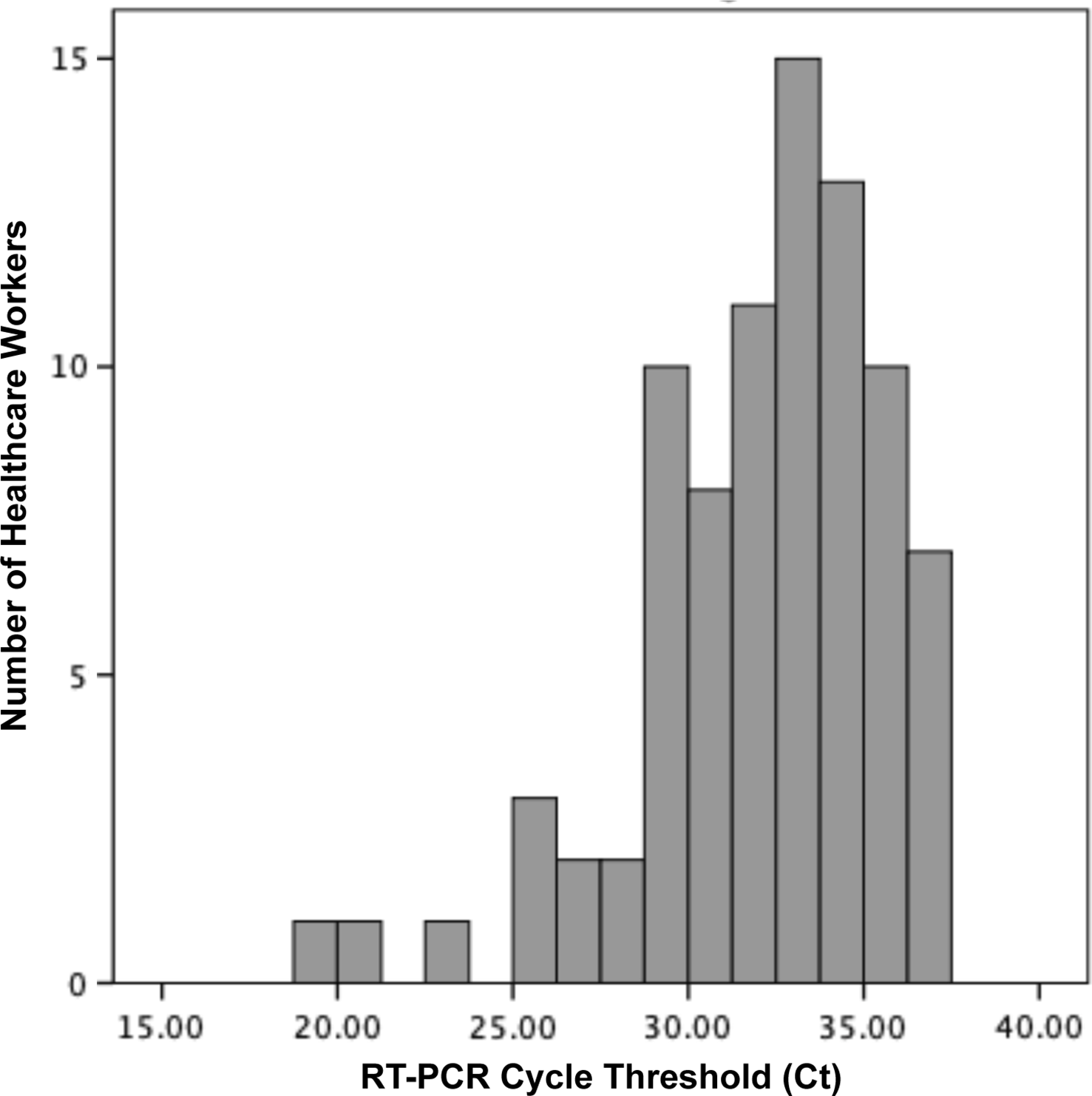
Fig. 1. Distribution of reverse-transcriptase polymerase chain reaction (RT-PCR) cycle threshold (Ct) values of 84 healthcare workers with confirmed COVID-19 and persistently positive respiratory sample for SARS-CoV-2 by RT-PCR, 14 days after the onset of symptoms.
Characteristics of positive healthcare worker
The HCW with the positive viral culture was a 43-year-old female beneficiary attendant. She presented a mild illness that included sore throat, cough, fever, myalgia, and diarrhea. She did not receive steroids or hydroxychloroquine, and she was not hospitalized. On the day of her sampling (day 16 of her illness), she had no symptoms other than a mild residual cough. None of her household contacts developed COVID-19. She remained positive by RT-PCR until at least 51 days after symptom onset, at which moment serial testing was stopped.
Discussion
We report the resolution of infectivity in most, but not all, HCWs with confirmed COVID-19 infection ∼2 weeks after symptom onset. Shedding of infectious virions may be prolonged in a very small subset of HCWs. Some jurisdictions allow a return to work after 10 days of illness. The low rate of infectivity 2 weeks after symptom onset highlights the need to enforce universal mask use and social distancing in healthcare settings because rare individuals may shed the virus for extended periods.
Few studies have investigated duration of infectivity. A German study of 9 patients with COVID-19 detected no viable virus by culture from respiratory specimens collected >8 days after symptom onset.Reference Wolfel, Corman and Guggemos4 In a second study of 47 patients in the United States, 1 of 5 specimens collected ≥8 days after symptom onset had infectious viruses detected by culture.Reference Arons, Hatfield and Reddy3 In Spain, viral culture was positive in 1 of 26 specimens collected ≥8 days post symptom onset (at day 10) in ambulatory patients.Reference Folgueira, Luczkowiak, Lasala, Perez-Rivilla and Delgado10 In Canada, viral cultures using a 4-day incubation period on samples previously frozen at −80°C were positive in 26 of the 72 specimens collected ≤7 days after symptom onset but were positive in 0 of 18 of those collected between days 8 and 11,Reference Bullard, Dust and Funk5 leading the authors to conclude that infectivity is limited to 8 days.Reference Bullard, Dust and Funk5 Our results suggest that longer periods of infectivity may occur. We did not identify risk factors for prolonged infectivity in the culture-positive HCW. Prolonged infectivity was also reported from a patient in Taiwan in whom virus was detected by culture 18 days after symptom onset.Reference Liu, Chang and Wang6 Infective virus can be found in specimens collected ≥3 weeks after symptom onset in hospitalized patients.Reference Folgueira, Luczkowiak, Lasala, Perez-Rivilla and Delgado10
Our study has strengths. To our knowledge, our study is the largest thus far among HCWs 2 weeks after onset of symptoms. We maximized the likelihood of detecting infective virus by avoiding freezing of specimens, prolonging the incubation period, and performing a final RT-PCR on supernatants. Our study has limitations. Even though most HCWs were sampled on day 14, some were sampled later. Our results cannot be generalized to individuals who may not be efficient to clear the virus (eg, the immunocompromised or elderly).
In conclusion, we report loss of viral infectivity in almost all HCWs whose symptoms have resolved 2 weeks after symptom onset. These findings should be considered when developing return-to-work policies.
Acknowledgments
Financial support
This work was supported by the Ministère de la santé et des services sociaux du Québec and the Fonds de recherche en santé du Québec [Y.L. and Q.C.]. These sponsors had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Conflicts of interest
Dr Longtin reports receiving research funding from Becton Dickinson and Merck. Dr Savard reports receiving honoraria from AVIR Pharma, Sunovion, and Verity Pharmaceuticals outside the submitted work. All other authors report no conflicts of interest relevant to this article.



