Studies of interventions to decrease the incidence of surgical site infections (SSIs) must include thousands of patients to achieve sufficient statistical power because SSI rates are typically low.Reference Schweizer, Cullen, Perencevich and Vaughan Sarrazin1 Traditional SSI detection relies on manual chart reviews, which is time-consuming and prone to subjective error and facility-level variation. Therefore, it is important to utilize data available from the electronic medical record (EMR) to accurately identify SSIs and to decrease the burden of manual chart reviews.Reference Pindyck, Gupta and Strymish2
Semiautomated or fully automated algorithms are 2 strategies for using EMR data for SSI detection.Reference van Mourik, Perencevich, Gastmeier and Bonten3 A semiautomated algorithm classifies cases as high or low likelihood of a SSI after it has occurred based on EMR data (eg, antibiotic administration) and prioritizes the time-intensive manual review process based on algorithm output. This strategy aims to maintain high sensitivity to detect all cases of SSIs while decreasing the burden of chart reviews, and, therefore, it is suitable for surveillance. Prior studies have reported significant workload reduction by developing semiautomated SSI detection algorithms.Reference Branch-Elliman, Strymish, Itani and Gupta4 Fully automated algorithms use EMR data to identify SSIs, without the added expense of time-intensive manual chart review. Due to the lack of confirmatory review, a fully automated algorithm aims to achieve a satisfactory positive predictive value (PPV) and specificity to identify SSIs, and it is suitable for research purposes.Reference van Mourik, Perencevich, Gastmeier and Bonten3
We aimed to develop a fully automated algorithm using data from the Veterans’ Affairs (VA) EMR to identify deep-incisional SSIs (ie, SSIs involving deep soft tissues of the incision, such as fascial and muscle layers) or organ-space SSIs (ie, SSIs involving any part of the body deeper than the fascial or muscle layers that was opened or manipulated during the operative procedure) after cardiac or orthopedic surgery.5
Methods
We conducted a retrospective cohort study of patients who underwent 1 of 3 primary surgeries: coronary artery bypass grafting, valve replacement, or total joint arthroplasty (TJA, which indicates total hip arthroplasty or total knee arthroplasty) at 11 VA hospitals. The TJA cohort was derived from data collected between January 1, 2007, and March 31, 2018, and the cardiac surgery cohort was derived from data collected between January 1, 2010, and March 31, 2018. Data collection for the cardiac surgery cohort began in 2010 due to changes in the way the VA measured cardiac surgery outcomes before that date. Data from the VA’s integrated EMR were obtained from the Corporate Data Warehouse (CDW) through the Veterans’ Affairs Informatics and Computing Infrastructure (VINCI).
Data were collected for positive microbiology results, erythrocyte sedimentation rate (ESR), C-reactive protein (CRP), type and duration of inpatient antibiotic orders and outpatient oral antibiotic orders prescribed, length of stay at admission for primary surgery, readmission, consultation notes, and administrative code data (ACD) including International Classification of Disease, Ninth or Tenth Revision (ICD-9 or -10) codes for SSIs and current procedural terminology (CPT) codes for reoperation for an infection-related purpose. Consultation notes were extracted as text fields, and we conducted a search for SSI-related key words (Supplemental Table 1 online). Positive microbiology results were collected for blood cultures and local cultures and categorized as Staphylococcus aureus, coagulase-negative Staphylococcus spp, gram-positive cocci other than Staphylococcus spp, gram-negative rods, and other organisms. The highest value of CRP and ESR during follow-up period was recorded and used for analyses. ICD codes and CPT codes were selected to capture possible SSI diagnoses broadly, using codes described in previous studies.Reference Yokoe, Avery, Platt and Huang6,Reference Huang, Placzek and Livingston7
Table 1. Bivariate and Multivariate Analysis of Variables Associated With Surgical Site Infection

Note. OR, odds ratio; CI, confidence interval; SSI, surgical site infection; ICD, International Classification of Diseases; MRSA, methicillin-resistant Staphylococcus aureus; ESR, erythrocyte sedimentation rate; CRP, C-reactive protein; GPC, gram-positive cocci; GNR, gram-negative rod.
The outcome of interest was mediastinitis, endocarditis, or deep-incisional or organ-space SSI within 30 days after surgery. The algorithm was tested for accuracy through comparison with outcomes collected via the Veterans’ Affairs Surgical Quality Improvement Program (VASQIP).Reference Massarweh, Kaji and Itani8 VASQIP is a national program that uses a validated sampling algorithm to direct manual review for SSI surveillance, and it is very similar to the National Surgical Quality Improvement Program. VASQIP applies National Healthcare Safety Network (NHSN) definitions, with the exception of a 30-day end point for all types of SSI.Reference Henderson and Daley9 However, VASQIP only reviews 70% of all major surgical cases. Patients without VASQIP review were excluded from the analysis. We elected to not to include superficial SSIs because superficial SSIs are subject to subjective bias and VASQIP did not collect superficial SSIs for large duration of study period.
We used multiple logistic regression analysis with repeated regular bootstrap procedure to select model variables. To build a model to predict SSIs, we started by assessing each candidate variable for bivariable associations with outcomes. We fit a multivariable logistic regression with SSIs as the outcome. To avoid overfitting of the model to the data, we selected variables using a backward elimination strategy to minimize Akaike’s information criterion (AIC). This variable selection was repeated 1,000 times using a bootstrapping method proposed by Austin and TuReference Austin and Tu10 to select only those variables identified as predictors in at least 90% of the bootstrap samples.
We then estimated coefficients of this multivariable logistic regression model by maximum likelihood estimation and used them to assign points for included variables. Coefficient values were approximated to the nearest integers for the ease of calculation and to improve practicality of the algorithm. We assigned points based on the created algorithm, and we measured sensitivity, specificity, PPV and negative predictive value (NPV) for each cutoff. We also used these summary statistics at each cutoff to create a receiver operating characteristic (ROC) curve to demonstrate the algorithm performance to predict SSIs. All statistical analyses were performed with SAS version 9.4 software (SAS Institution, Cary, NC).
The study was approved by the VA Central Institutional Review Board.
Results
Among the 11 VA hospitals, we identified 13,341 cardiac surgeries and 18,914 TJAs during the study period. Of these, 9,557 (72.0%) cardiac surgeries and 12,992 (69.0%) TJAs were evaluated by VASQIP and were included in our final cohort. Moreover, 49 (0.5%) cardiac surgeries were classified mediastinitis or endocarditis in VASQIP, and 83 TJAs (0.6%) were classified as deep-incisional or organ-space SSIs.
The ICD codes for infection, subsequent surgery (sternal debridement in cardiac surgeries and reoperation in TJAs) and positive cultures for S. aureus were included in both models (Table 1). Sensitivities, specificities, PPVs and NPVs in varying cutoffs are shown in Table 2. With ≥60% sensitivity, the PPV of the SSI detection algorithm after cardiac surgeries was 52.5%. With 80% sensitivity, the PPV of the SSI detection algorithm after TJA was 62.0%. The area under the curve (AUC) of the receiver operating characteristic (ROC) curve for SSI detection algorithms after cardiac surgeries and TJAs were 0.96 and 0.97 in the final models, respectively (Fig. 1).

Fig. 1. Receiver operating characteristic (ROC) curve for surgical site infection (SSI) detection algorithms.
Table 2. Sensitivities, Specificities, PPV Positive Predictive Values and NPV Negative Predictive Values of Surgical Site Infection Detection Algorithms, With Varying Cutoffs
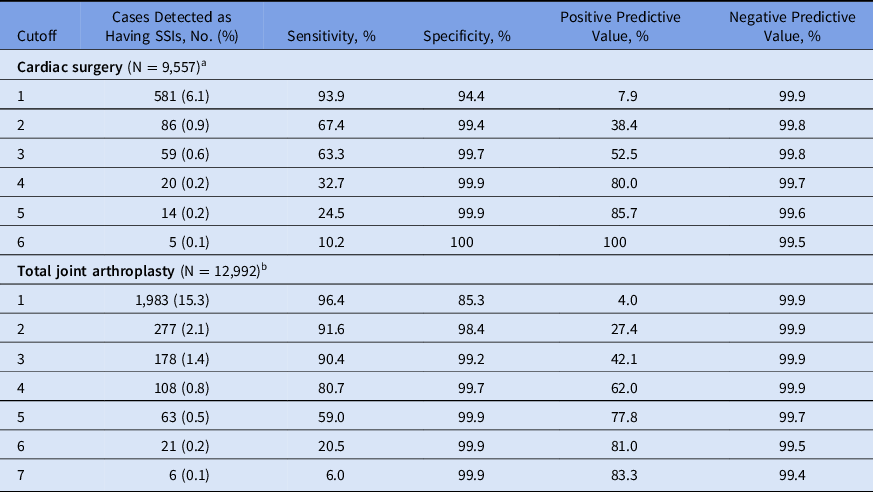
Note. PPV, positive predictive values; NPV, negative predictive values; SSI, surgical site infection; ESR, erythrocyte sedimentation rate; ICD, International Classification of Diseases.
a Positive culture for Staphylococcus aureus = 1 points, sternal debridement = 2 points, ICD codes for other SSIs = 2 points, ICD codes for mediastinitis = 1 point.
b ICD codes for definite SSIs = 2 points, ICD codes for other diagnosis for possible SSIs = 2 points, any consultation note mentioning hip or knee surgical infection = 1 point, reoperation = 1 point, readmission = 1 point, positive culture for Staphylococcus aureus = 1 point, use of vancomycin or piperacillin/tazobactam ≥6 days = 1 point, use of rifampin = 1 point.
Discussion
We used data from the existing VA EMR to develop a fully automated SSI detection algorithms for cardiac surgeries and TJAs. With VASQIP as a reference standard, the algorithms achieved high specificities and NPVs, with PPVs of 52.5% for cardiac surgeries and 62.0% for TJAs while maintaining at least 60% sensitivity. Considering the low prevalence rate of SSIs, our algorithms were successful in identifying a majority of patients with a true SSI while simultaneously reducing false-positive cases.
Our study utilized detailed clinical data combined with ACD to differentiate cases with and without SSIs. ACD are widely used in surveillance and public reporting programs but are insufficient as a sole indicator for detecting SSIs.Reference Goto, Ohl, Schweizer and Perencevich11 In addition to ACD, positive clinical cultures for S. aureus were assigned high points. The possible rationale behind this is that S. aureus is the most common pathogen causing postoperative mediastinitis and early-onset prosthetic joint infections. In our study, the performance of the SSI detection algorithm for TJAs was better than that for cardiac surgeries. This difference is probably due to the better performance of ACD used for SSIs after orthopedic surgeries, compared to those used for SSIs after cardiac surgeries.Reference Goto, Ohl, Schweizer and Perencevich11,Reference Bolon, Hooper and Stevenson12
This study has several limitations. First, we only evaluated patients for 30 days after index surgery because VASQIP only detects SSIs within 30 days. This method differs from NHSN surveillance that detects SSIs within 90 days for TJA and cardiac surgeries. We might have missed SSIs caused by more indolent organisms with onset beyond the 30-day period. Second, only deep-incisional and organ-space SSIs were targeted in our algorithm because the symptoms and definitions associated with superficial infections are vague. Third, different ACD (ICD-9/-10) were used in different time frames during study period. ICD-9/-10 translations do not always align and produce discontinuity in some diagnoses.Reference Mainor, Morden, Smith, Tomlin and Skinner13 Fourth, we evaluated only positive clinical cultures; thus, we might have missed some culture-negative SSIs. However, this limitation was mitigated by other elements of the algorithm, which would have flagged patients in other ways, such as ACD or antimicrobial orders. Finally, due to limitations of the EMR, we could not obtain outpatient intravenous antibiotic data; however, the vast majority of patients with deep-incisional or organ-space SSIs are first evaluated on an inpatient basis; thus, those orders would have been included in the algorithm.
Future research to improve these algorithms should include machine-learning approaches or advanced key word searches for infection-related terms, as has been applied for measurement of infections following cardiac device procedures within the VA.Reference Mull, Stolzmann, Shin, Kalver, Schweizer and Branch-Elliman14 These algorithms should also be validated in other healthcare systems with EMR data using NHSN data as the gold standard.
Current research studies of interventions to prevent SSI rely on either ACD alone or on data from intensive chart review to assess whether the interventions were successful.Reference Goto, Ohl, Schweizer and Perencevich11,Reference Schweizer, Chiang and Septimus15 These algorithms will improve our ability to accurately evaluate SSI interventions in large healthcare systems. In the VA context, these algorithms can be combined with VASQIP data to assess SSIs among all relevant surgical patients in the VA system, rather than only the proportion of patients assessed by VASQIP.
Acknowledgments
The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of the Department of Veterans’ Affairs or the United States government.
Financial support
This study was funded by the VA Health Services Research and Development Service (grant no. CRE 12-291; primary investigator, Perencevich).
Conflicts of interest
Dr Westyn Branch-Elliman is supported by National Institutes of Health (grant no. NHLBI 1K12HL138049-01). All other authors report no conflicts of interest relevant to this article.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/ice.2020.1387






