Editorial
Editorial
-
- Published online by Cambridge University Press:
- 20 February 2008, p. i
-
- Article
-
- You have access
- HTML
- Export citation
Deborah Charlesworth
editorial
-
- Published online by Cambridge University Press:
- 20 February 2008, p. 1
-
- Article
-
- You have access
- HTML
- Export citation
Paper
Asymmetrical mating patterns and the evolution of biased style-morph ratios in a tristylous daffodil
-
- Published online by Cambridge University Press:
- 20 February 2008, pp. 3-15
-
- Article
-
- You have access
- HTML
- Export citation
The number, age, sharing and relatedness of S-locus specificities in Prunus
-
- Published online by Cambridge University Press:
- 20 February 2008, pp. 17-26
-
- Article
-
- You have access
- HTML
- Export citation
When gametophytic self-incompatibility meets gynodioecy
-
- Published online by Cambridge University Press:
- 20 February 2008, pp. 27-35
-
- Article
-
- You have access
- HTML
- Export citation
Hitch-hiking to a locus under balancing selection: high sequence diversity and low population subdivision at the S-locus genomic region in Arabidopsis halleri
-
- Published online by Cambridge University Press:
- 20 February 2008, pp. 37-46
-
- Article
-
- You have access
- HTML
- Export citation
Genetic causes and consequences of the breakdown of self-incompatibility: case studies in the Brassicaceae
-
- Published online by Cambridge University Press:
- 20 February 2008, pp. 47-60
-
- Article
-
- You have access
- HTML
- Export citation
Prior selfing and the selfing syndrome in animals: an experimental approach in the freshwater snail Biomphalaria pfeifferi
-
- Published online by Cambridge University Press:
- 20 February 2008, pp. 61-72
-
- Article
-
- You have access
- HTML
- Export citation
Consequences of inbreeding depression due to sex-linked loci for the maintenance of males and outcrossing in branchiopod crustaceans
-
- Published online by Cambridge University Press:
- 20 February 2008, pp. 73-84
-
- Article
-
- You have access
- HTML
- Export citation
A selective sweep in or near the Silene latifolia X-linked gene SlssX
-
- Published online by Cambridge University Press:
- 20 February 2008, pp. 85-95
-
- Article
-
- You have access
- HTML
- Export citation
Mating system and recombination affect molecular evolution in four Triticeae species
-
- Published online by Cambridge University Press:
- 20 February 2008, pp. 97-109
-
- Article
-
- You have access
- HTML
- Export citation
‘Junk’ DNA and phenotypic evolution in Silene section Siphonomorpha
-
- Published online by Cambridge University Press:
- 20 February 2008, pp. 111-118
-
- Article
-
- You have access
- HTML
- Export citation
Effective population size and tests of neutrality at cytoplasmic genes in Arabidopsis
-
- Published online by Cambridge University Press:
- 20 February 2008, pp. 119-128
-
- Article
-
- You have access
- HTML
- Export citation
A Bayesian method for jointly estimating allele age and selection intensity†
-
- Published online by Cambridge University Press:
- 20 February 2008, pp. 129-137
-
- Article
-
- You have access
- HTML
- Export citation
The effect of a barrier to gene flow on patterns of geographic variation
-
- Published online by Cambridge University Press:
- 20 February 2008, pp. 139-149
-
- Article
-
- You have access
- HTML
- Export citation


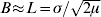 ) are significant in one dimension (μ is the rate of mutation or long-range dispersal). Thus, in a two-dimensional population, barriers to gene flow can be detected through their effect on the spatial pattern of genetic marker alleles.
) are significant in one dimension (μ is the rate of mutation or long-range dispersal). Thus, in a two-dimensional population, barriers to gene flow can be detected through their effect on the spatial pattern of genetic marker alleles.