Published online by Cambridge University Press: 14 April 2009
Genetic variation contained in a multigene family was theoretically investigated from the standpoint of population genetics. Unequal crossover is assumed to be responsible for the coincidental evolution of mutant genes in a chromosome. When the allowed latitude of the duplicated or deleted number of gene units at unequal crossover is 10 ˜ 15% of the total gene number in a chromosome, the arrangement of gene lineage in a chromosome is shown to be roughly random. The equilibrium properties of genetic variation or the probability of identity of two genes within a family (clonality) were studied under mutation, unequal crossover, interchromosomal crossover and sampling of gametes. The clonality of a multigene family within a chromosome is shown to be approximately
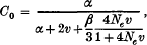
in which α = 2k/n2 with k = effective number of cycles of unequal crossover and with n = number of gene units in a family, v is the mutation rate per gene unit, β is the rate of interchromosomal crossover per family and Ne is the effective size of the population, all measured by the rate per generation. The clonality of a gene family between two different chromosomes becomes approximately C1 = C0/(l + 4Neν). Some models of natural selection which lowers the clonality or increases genetic variation in a multigene family were investigated. It was shown that natural selection may be quite effective in increasing genetic variation in a gene family.